Structure of the primed paramyxovirus fusion protein
- PMID: 23027936
- PMCID: PMC3478655
- DOI: 10.1073/pnas.1214903109
Structure of the primed paramyxovirus fusion protein
Conflict of interest statement
The authors declare no conflict of interest.
Figures
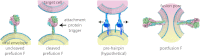
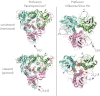
Comment on
-
Structure of the cleavage-activated prefusion form of the parainfluenza virus 5 fusion protein.Proc Natl Acad Sci U S A. 2012 Oct 9;109(41):16672-7. doi: 10.1073/pnas.1213802109. Epub 2012 Sep 10. Proc Natl Acad Sci U S A. 2012. PMID: 23012473 Free PMC article.
References
-
- Skehel JJ, Wiley DC. Coiled coils in both intracellular vesicle and viral membrane fusion. Cell. 1998;95:871–874. - PubMed
-
- Bizebard T, et al. Structure of influenza virus haemagglutinin complexed with a neutralizing antibody. Nature. 1995;376:92–94. - PubMed
-
- Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

