In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters
- PMID: 23027949
- PMCID: PMC3479502
- DOI: 10.1073/pnas.1211759109
In vitro reconstitution of the ordered assembly of the endosomal sorting complex required for transport at membrane-bound HIV-1 Gag clusters
Abstract
Most membrane-enveloped viruses depend on host proteins of the endosomal sorting complex required for transport (ESCRT) machinery for their release. HIV-1 is the prototypic ESCRT-dependent virus. The direct interactions between HIV-1 and the early ESCRT factors TSG101 and ALIX have been mapped in detail. However, the full pathway of ESCRT recruitment to HIV-1 budding sites, which culminates with the assembly of the late-acting CHMP4, CHMP3, CHMP2, and CHMP1 subunits, is less completely understood. Here, we report the biochemical reconstitution of ESCRT recruitment to viral assembly sites, using purified proteins and giant unilamellar vesicles. The myristylated full-length Gag protein of HIV-1 was purified to monodispersity. Myr-Gag forms clusters on giant unilamellar vesicle membranes containing the plasma membrane lipid PI(4,5)P(2). These Gag clusters package a fluorescent oligonucleotide, and recruit early ESCRT complexes ESCRT-I or ALIX with the appropriate dependence on the Gag PTAP and LYP(X)(n)L motifs. ALIX directly recruits the key ESCRT-III subunit CHMP4. ESCRT-I can only recruit CHMP4 when ESCRT-II and CHMP6 are present as intermediary factors. Downstream of CHMP4, CHMP3 and CHMP2 assemble synergistically, with the presence of both subunits required for efficient recruitment. The very late-acting factor CHMP1 is not recruited unless the pathway is completed through CHMP3 and CHMP2. These findings define the minimal sets of components needed to complete ESCRT assembly at HIV-1 budding sites, and provide a starting point for in vitro structural and biophysical dissection of the system.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

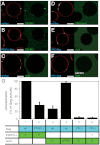
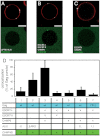
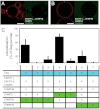

References
-
- Martin-Serrano J, Neil SJD. Host factors involved in retroviral budding and release. Nat Rev Microbiol. 2011;9:519–531. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

