Using intein catalysis to probe the origin of major histocompatibility complex class I-presented peptides
- PMID: 23027972
- PMCID: PMC3479494
- DOI: 10.1073/pnas.1210271109
Using intein catalysis to probe the origin of major histocompatibility complex class I-presented peptides
Abstract
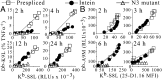
All vertebrate nucleated cells generate peptides from their expressed gene products and then display them at the cell surface bound to MHC class I molecules. This allows CD8(+) T cells to detect and eliminate abnormal cells that are synthesizing foreign proteins, e.g., from viruses or mutations. To permit the immune system to more uniformly monitor a cell's proteins, regardless of their half-life or location, it has been thought that the products of rapid degradation of the mistakes of protein synthesis (defective ribosomal products, DRiPs) preferentially contribute to the class I-presented peptides. However, using intein catalysis to generate peptide sequences exclusively by posttranslational splicing of mature proteins, we show here that presented peptides can be generated from fully folded and functional proteins. Remarkably, the presentation of peptides from two model mature proteins is just as efficient as from newly synthesized proteins subject to errors in translation or folding. These results indicate that for the constructs we have analyzed, DRiPs are not a more efficient source of class I peptides for antigen presentation than the turnover of mature functional proteins. Accordingly, our data suggest that one of the major ways the immune system evaluates the health of cells is by monitoring the breakdown products of the proteome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


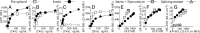

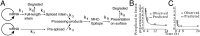
References
-
- Goldberg AL, Dice JF. Intracellular protein degradation in mammalian and bacterial cells. Annu Rev Biochem. 1974;43:835–869. - PubMed
-
- Hershko A, Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. - PubMed
-
- Rock KL, et al. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell. 1994;78:761–771. - PubMed
-
- Yewdell JW, Antón LC, Bennink JR. Defective ribosomal products (DRiPs): A major source of antigenic peptides for MHC class I molecules? J Immunol. 1996;157:1823–1826. - PubMed
-
- Boon T, Van Pel A. T cell-recognized antigenic peptides derived from the cellular genome are not protein degradation products but can be generated directly by transcription and translation of short subgenic regions. A hypothesis. Immunogenetics. 1989;29:75–79. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

