The RNA polymerase II CTD coordinates transcription and RNA processing
- PMID: 23028141
- PMCID: PMC3465734
- DOI: 10.1101/gad.200303.112
The RNA polymerase II CTD coordinates transcription and RNA processing
Abstract
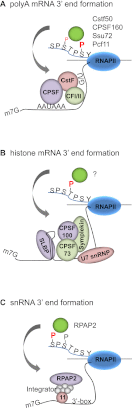
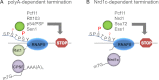
The C-terminal domain (CTD) of the RNA polymerase II largest subunit consists of multiple heptad repeats (consensus Tyr1-Ser2-Pro3-Thr4-Ser5-Pro6-Ser7), varying in number from 26 in yeast to 52 in vertebrates. The CTD functions to help couple transcription and processing of the nascent RNA and also plays roles in transcription elongation and termination. The CTD is subject to extensive post-translational modification, most notably phosphorylation, during the transcription cycle, which modulates its activities in the above processes. Therefore, understanding the nature of CTD modifications, including how they function and how they are regulated, is essential to understanding the mechanisms that control gene expression. While the significance of phosphorylation of Ser2 and Ser5 residues has been studied and appreciated for some time, several additional modifications have more recently been added to the CTD repertoire, and insight into their function has begun to emerge. Here, we review findings regarding modification and function of the CTD, highlighting the important role this unique domain plays in coordinating gene activity.
Figures



References
-
- Aguilera A, Garcia-Muse T 2012. R loops: From transcription byproducts to threats to genome stability. Mol Cell 46: 115–124 - PubMed
-
- Ahn SH, Kim M, Buratowski S 2004. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol Cell 13: 67–76 - PubMed
-
- Akoulitchev S, Makela TP, Weinberg RA, Reinberg D 1995. Requirement for TFIIH kinase activity in transcription by RNA polymerase II. Nature 377: 557–560 - PubMed
-
- Akoulitchev S, Chuikov S, Reinberg D 2000. TFIIH is negatively regulated by cdk8-containing mediator complexes. Nature 407: 102–106 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
