Promoter-proximal polyadenylation sites reduce transcription activity
- PMID: 23028143
- PMCID: PMC3465738
- DOI: 10.1101/gad.189126.112
Promoter-proximal polyadenylation sites reduce transcription activity
Abstract
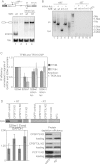
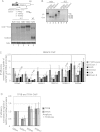
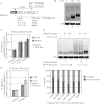
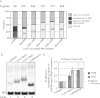
Gene expression relies on the functional communication between mRNA processing and transcription. We previously described the negative impact of a point-mutated splice donor (SD) site on transcription. Here we demonstrate that this mutation activates an upstream cryptic polyadenylation (CpA) site, which in turn causes reduced transcription. Functional depletion of U1 snRNP in the context of the wild-type SD triggers the same CpA event accompanied by decreased RNA levels. Thus, in accordance with recent findings, U1 snRNP can shield premature pA sites. The negative impact of unshielded pA sites on transcription requires promoter proximity, as demonstrated using artificial constructs and supported by a genome-wide data set. Importantly, transcription down-regulation can be recapitulated in a gene context devoid of splice sites by placing a functional bona fide pA site/transcription terminator within ~500 base pairs of the promoter. In contrast, promoter-proximal positioning of a pA site-independent histone gene terminator supports high transcription levels. We propose that optimal communication between a pA site-dependent gene terminator and its promoter critically depends on gene length and that short RNA polymerase II-transcribed genes use specialized termination mechanisms to maintain high transcription levels.
Figures


References
-
- Ashe MP, Griffin P, James W, Proudfoot NJ 1995. Poly(A) site selection in the HIV-1 provirus: Inhibition of promoter-proximal polyadenylation by the downstream major splice donor site. Genes Dev 9: 3008–3025 - PubMed
-
- Baillat D, Hakimi MA, Näär AM, Shilatifard A, Cooch N, Shiekhattar R 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repreat of RNA polymerase II. Cell 123: 265–276 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
