miR-26a is required for skeletal muscle differentiation and regeneration in mice
- PMID: 23028144
- PMCID: PMC3465739
- DOI: 10.1101/gad.198085.112
miR-26a is required for skeletal muscle differentiation and regeneration in mice
Abstract
Multiple microRNAs are known to be induced during the differentiation of myoblasts to myotubes. Yet, experiments in animals have not provided clear evidence for the requirement of most of these microRNAs in myogenic differentiation in vivo. miR-26a is induced during skeletal muscle differentiation and is predicted to target a well-known inhibitor of differentiation, the transforming growth factor β/bone morphogenetic protein (TGF-β/BMP) signaling pathway. Here we show that exogenous miR-26a promotes differentiation of myoblasts, while inhibition of miR-26a by antisense oligonucleotides or by Tough-Decoys delays differentiation. miR-26a targets the transcription factors Smad1 and Smad4, critical for the TGF-β/BMP pathway, and expression of microRNA-resistant forms of these transcription factors inhibits differentiation. Injection of antagomirs specific to miR-26a into neonatal mice derepressed both Smad expression and activity and consequently inhibited skeletal muscle differentiation. In addition, miR-26a is induced during skeletal muscle regeneration after injury. Inhibiting miR-26a in the tibialis anterior muscles through the injection of adeno-associated virus expressing a Tough-Decoy targeting miR-26a prevents Smad down-regulation and delays regeneration. These findings provide evidence for the requirement of miR-26a for skeletal muscle differentiation and regeneration in vivo.
Figures



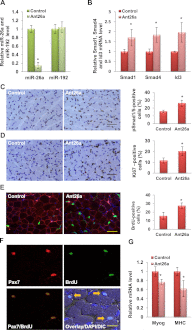

Similar articles
-
miR-431 promotes differentiation and regeneration of old skeletal muscle by targeting Smad4.Genes Dev. 2015 Aug 1;29(15):1605-17. doi: 10.1101/gad.263574.115. Epub 2015 Jul 27. Genes Dev. 2015. PMID: 26215566 Free PMC article.
-
microRNA-1 and microRNA-206 regulate skeletal muscle satellite cell proliferation and differentiation by repressing Pax7.J Cell Biol. 2010 Sep 6;190(5):867-79. doi: 10.1083/jcb.200911036. J Cell Biol. 2010. PMID: 20819939 Free PMC article.
-
The H19 long noncoding RNA gives rise to microRNAs miR-675-3p and miR-675-5p to promote skeletal muscle differentiation and regeneration.Genes Dev. 2014 Mar 1;28(5):491-501. doi: 10.1101/gad.234419.113. Epub 2014 Feb 14. Genes Dev. 2014. PMID: 24532688 Free PMC article.
-
Effects of microRNAs on skeletal muscle development.Gene. 2018 Aug 20;668:107-113. doi: 10.1016/j.gene.2018.05.039. Epub 2018 May 25. Gene. 2018. PMID: 29775754 Review.
-
MicroRNAs involved in skeletal muscle differentiation.J Genet Genomics. 2013 Mar 20;40(3):107-16. doi: 10.1016/j.jgg.2013.02.002. Epub 2013 Feb 20. J Genet Genomics. 2013. PMID: 23522383 Review.
Cited by
-
MiR-26a inhibits stem cell-like phenotype and tumor growth of osteosarcoma by targeting Jagged1.Oncogene. 2017 Jan 12;36(2):231-241. doi: 10.1038/onc.2016.194. Epub 2016 Jun 6. Oncogene. 2017. PMID: 27270422
-
Characteristics of microRNAs in Skeletal Muscle of Intrauterine Growth-Restricted Pigs.Genes (Basel). 2023 Jun 28;14(7):1372. doi: 10.3390/genes14071372. Genes (Basel). 2023. PMID: 37510277 Free PMC article.
-
Integrative analysis of porcine microRNAome during skeletal muscle development.PLoS One. 2013 Sep 11;8(9):e72418. doi: 10.1371/journal.pone.0072418. eCollection 2013. PLoS One. 2013. PMID: 24039761 Free PMC article.
-
A Novel Method for Detecting Duchenne Muscular Dystrophy in Blood Serum of mdx Mice.Genes (Basel). 2022 Jul 27;13(8):1342. doi: 10.3390/genes13081342. Genes (Basel). 2022. PMID: 36011258 Free PMC article.
-
MicroRNA-542-3p targets Pten to inhibit the myoblasts proliferation but suppresses myogenic differentiation independent of targeted Pten.BMC Genomics. 2024 Apr 1;25(1):325. doi: 10.1186/s12864-024-10260-y. BMC Genomics. 2024. PMID: 38561670 Free PMC article.
References
-
- Bartel DP 2004. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 116: 281–297 - PubMed
-
- Berkes CA, Tapscott SJ 2005. MyoD and the transcriptional control of myogenesis. Semin Cell Dev Biol 16: 585–595 - PubMed
-
- Buckingham M, Bajard L, Daubas P, Esner M, Lagha M, Relaix F, Rocancourt D 2006. Myogenic progenitor cells in the mouse embryo are marked by the expression of Pax3/7 genes that regulate their survival and myogenic potential. Anat Embryol 211: 51–56 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
