Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks
- PMID: 23028288
- PMCID: PMC3459874
- DOI: 10.1371/journal.pcbi.1002690
Enhancing the prioritization of disease-causing genes through tissue specific protein interaction networks
Abstract
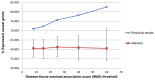
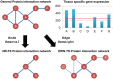
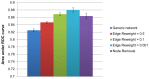
The prioritization of candidate disease-causing genes is a fundamental challenge in the post-genomic era. Current state of the art methods exploit a protein-protein interaction (PPI) network for this task. They are based on the observation that genes causing phenotypically-similar diseases tend to lie close to one another in a PPI network. However, to date, these methods have used a static picture of human PPIs, while diseases impact specific tissues in which the PPI networks may be dramatically different. Here, for the first time, we perform a large-scale assessment of the contribution of tissue-specific information to gene prioritization. By integrating tissue-specific gene expression data with PPI information, we construct tissue-specific PPI networks for 60 tissues and investigate their prioritization power. We find that tissue-specific PPI networks considerably improve the prioritization results compared to those obtained using a generic PPI network. Furthermore, they allow predicting novel disease-tissue associations, pointing to sub-clinical tissue effects that may escape early detection.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Birnbaum S, Ludwig KU, Reutter H, Herms S, Steffens M, et al. (2009) Key susceptibility locus for nonsyndromic cleft lip with or without cleft palate on chromosome 8q24. Nat Genet 41: 473–477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

