Tissue-specific functional networks for prioritizing phenotype and disease genes
- PMID: 23028291
- PMCID: PMC3459891
- DOI: 10.1371/journal.pcbi.1002694
Tissue-specific functional networks for prioritizing phenotype and disease genes
Abstract
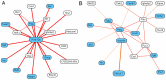
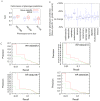
Integrated analyses of functional genomics data have enormous potential for identifying phenotype-associated genes. Tissue-specificity is an important aspect of many genetic diseases, reflecting the potentially different roles of proteins and pathways in diverse cell lineages. Accounting for tissue specificity in global integration of functional genomics data is challenging, as "functionality" and "functional relationships" are often not resolved for specific tissue types. We address this challenge by generating tissue-specific functional networks, which can effectively represent the diversity of protein function for more accurate identification of phenotype-associated genes in the laboratory mouse. Specifically, we created 107 tissue-specific functional relationship networks through integration of genomic data utilizing knowledge of tissue-specific gene expression patterns. Cross-network comparison revealed significantly changed genes enriched for functions related to specific tissue development. We then utilized these tissue-specific networks to predict genes associated with different phenotypes. Our results demonstrate that prediction performance is significantly improved through using the tissue-specific networks as compared to the global functional network. We used a testis-specific functional relationship network to predict genes associated with male fertility and spermatogenesis phenotypes, and experimentally confirmed one top prediction, Mbyl1. We then focused on a less-common genetic disease, ataxia, and identified candidates uniquely predicted by the cerebellum network, which are supported by both literature and experimental evidence. Our systems-level, tissue-specific scheme advances over traditional global integration and analyses and establishes a prototype to address the tissue-specific effects of genetic perturbations, diseases and drugs.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

