A novel rhabdovirus associated with acute hemorrhagic fever in central Africa
- PMID: 23028323
- PMCID: PMC3460624
- DOI: 10.1371/journal.ppat.1002924
A novel rhabdovirus associated with acute hemorrhagic fever in central Africa
Erratum in
-
Correction: A Novel Rhabdovirus Associated with Acute Hemorrhagic Fever in Central Africa.PLoS Pathog. 2016 Mar 18;12(3):e1005503. doi: 10.1371/journal.ppat.1005503. eCollection 2016 Mar. PLoS Pathog. 2016. PMID: 26991269 Free PMC article. No abstract available.
-
Correction: Correction: A Novel Rhabdovirus Associated with Acute Hemorrhagic Fever in Central Africa.PLoS Pathog. 2017 Sep 7;13(9):e1006583. doi: 10.1371/journal.ppat.1006583. eCollection 2017 Sep. PLoS Pathog. 2017. PMID: 28880960 Free PMC article.
Abstract
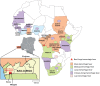
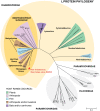
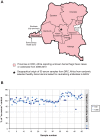
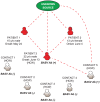
Deep sequencing was used to discover a novel rhabdovirus (Bas-Congo virus, or BASV) associated with a 2009 outbreak of 3 human cases of acute hemorrhagic fever in Mangala village, Democratic Republic of Congo (DRC), Africa. The cases, presenting over a 3-week period, were characterized by abrupt disease onset, high fever, mucosal hemorrhage, and, in two patients, death within 3 days. BASV was detected in an acute serum sample from the lone survivor at a concentration of 1.09 × 10(6) RNA copies/mL, and 98.2% of the genome was subsequently de novo assembled from ≈ 140 million sequence reads. Phylogenetic analysis revealed that BASV is highly divergent and shares less than 34% amino acid identity with any other rhabdovirus. High convalescent neutralizing antibody titers of >1:1000 were detected in the survivor and an asymptomatic nurse directly caring for him, both of whom were health care workers, suggesting the potential for human-to-human transmission of BASV. The natural animal reservoir host or arthropod vector and precise mode of transmission for the virus remain unclear. BASV is an emerging human pathogen associated with acute hemorrhagic fever in Africa.
Conflict of interest statement
The authors have filed a patent application related to BASV. This does not alter the authors' adherence to all PLOS Pathogens policies on sharing data and materials.
Figures




References
-
- Bray M (2005) Pathogenesis of viral hemorrhagic fever. Curr Opin Immunol 17: 399–403. - PubMed
-
- Carrion R Jr, Patterson JL (2011) Vaccines against viral hemorrhagic fevers: non-human primate models. Hum Vaccin 7: 667–673. - PubMed
-
- Geisbert TW, Jahrling PB (2004) Exotic emerging viral diseases: progress and challenges. Nat Med 10: S110–121. - PubMed
-
- Borio L, Inglesby T, Peters CJ, Schmaljohn AL, Hughes JM, et al. (2002) Hemorrhagic fever viruses as biological weapons: medical and public health management. JAMA 287: 2391–2405. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

