Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast
- PMID: 23028377
- PMCID: PMC3447972
- DOI: 10.1371/journal.pgen.1002985
Factors that promote H3 chromatin integrity during transcription prevent promiscuous deposition of CENP-A(Cnp1) in fission yeast
Abstract
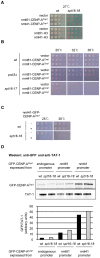
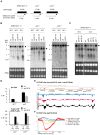
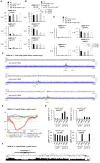
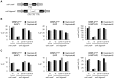
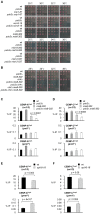
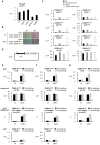
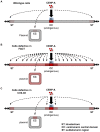
Specialized chromatin containing CENP-A nucleosomes instead of H3 nucleosomes is found at all centromeres. However, the mechanisms that specify the locations at which CENP-A chromatin is assembled remain elusive in organisms with regional, epigenetically regulated centromeres. It is known that normal centromeric DNA is transcribed in several systems including the fission yeast, Schizosaccharomyces pombe. Here, we show that factors which preserve stable histone H3 chromatin during transcription also play a role in preventing promiscuous CENP-A(Cnp1) deposition in fission yeast. Mutations in the histone chaperone FACT impair the maintenance of H3 chromatin on transcribed regions and promote widespread CENP-A(Cnp1) incorporation at non-centromeric sites. FACT has little or no effect on CENP-A(Cnp1) assembly at endogenous centromeres where CENP-A(Cnp1) is normally assembled. In contrast, Clr6 complex II (Clr6-CII; equivalent to Rpd3S) histone deacetylase function has a more subtle impact on the stability of transcribed H3 chromatin and acts to prevent the ectopic accumulation of CENP-A(Cnp1) at specific loci, including subtelomeric regions, where CENP-A(Cnp1) is preferentially assembled. Moreover, defective Clr6-CII function allows the de novo assembly of CENP-A(Cnp1) chromatin on centromeric DNA, bypassing the normal requirement for heterochromatin. Thus, our analyses show that alterations in the process of chromatin assembly during transcription can destabilize H3 nucleosomes and thereby allow CENP-A(Cnp1) to assemble in its place. We propose that normal centromeres provide a specific chromatin context that limits reassembly of H3 chromatin during transcription and thereby promotes the establishment of CENP-A(Cnp1) chromatin and associated kinetochores. These findings have important implications for genetic and epigenetic processes involved in centromere specification.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

