Mining the Sinorhizobium meliloti transportome to develop FRET biosensors for sugars, dicarboxylates and cyclic polyols
- PMID: 23028462
- PMCID: PMC3454389
- DOI: 10.1371/journal.pone.0043578
Mining the Sinorhizobium meliloti transportome to develop FRET biosensors for sugars, dicarboxylates and cyclic polyols
Abstract
Background: Förster resonance energy transfer (FRET) biosensors are powerful tools to detect biologically important ligands in real time. Currently FRET bisosensors are available for twenty-two compounds distributed in eight classes of chemicals (two pentoses, two hexoses, two disaccharides, four amino acids, one nucleobase, two nucleotides, six ions and three phytoestrogens). To expand the number of available FRET biosensors we used the induction profile of the Sinorhizobium meliloti transportome to systematically screen for new FRET biosensors.
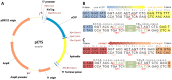
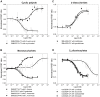
Methodology/principal findings: Two new vectors were developed for cloning genes for solute-binding proteins (SBPs) between those encoding FRET partner fluorescent proteins. In addition to a vector with the widely used cyan and yellow fluorescent protein FRET partners, we developed a vector using orange (mOrange2) and red fluorescent protein (mKate2) FRET partners. From the sixty-nine SBPs tested, seven gave a detectable FRET signal change on binding substrate, resulting in biosensors for D-quinic acid, myo-inositol, L-rhamnose, L-fucose, β-diglucosides (cellobiose and gentiobiose), D-galactose and C4-dicarboxylates (malate, succinate, oxaloacetate and fumarate). To our knowledge, we describe the first two FRET biosensor constructs based on SBPs from Tripartite ATP-independent periplasmic (TRAP) transport systems.
Conclusions/significance: FRET based on orange (mOrange2) and red fluorescent protein (mKate2) partners allows the use of longer wavelength light, enabling deeper penetration of samples at lower energy and increased resolution with reduced back-ground auto-fluorescence. The FRET biosensors described in this paper for four new classes of compounds; (i) cyclic polyols, (ii) L-deoxy sugars, (iii) β-linked disaccharides and (iv) C4-dicarboxylates could be developed to study metabolism in vivo.
Conflict of interest statement
Figures
Similar articles
-
Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB, and dctD.Mol Plant Microbe Interact. 1990 May-Jun;3(3):174-81. doi: 10.1094/mpmi-3-174. Mol Plant Microbe Interact. 1990. PMID: 2134335
-
Booster, a Red-Shifted Genetically Encoded Förster Resonance Energy Transfer (FRET) Biosensor Compatible with Cyan Fluorescent Protein/Yellow Fluorescent Protein-Based FRET Biosensors and Blue Light-Responsive Optogenetic Tools.ACS Sens. 2020 Mar 27;5(3):719-730. doi: 10.1021/acssensors.9b01941. Epub 2020 Feb 26. ACS Sens. 2020. PMID: 32101394
-
TRAP transporters: a new family of periplasmic solute transport systems encoded by the dctPQM genes of Rhodobacter capsulatus and by homologs in diverse gram-negative bacteria.J Bacteriol. 1997 Sep;179(17):5482-93. doi: 10.1128/jb.179.17.5482-5493.1997. J Bacteriol. 1997. PMID: 9287004 Free PMC article.
-
FÖrster resonance energy transfer (FRET)-based biosensors for biological applications.Biosens Bioelectron. 2019 Aug 1;138:111314. doi: 10.1016/j.bios.2019.05.019. Epub 2019 May 10. Biosens Bioelectron. 2019. PMID: 31096114 Review.
-
Two Decades of Genetically Encoded Biosensors Based on Förster Resonance Energy Transfer.Cell Struct Funct. 2019 Nov 2;44(2):153-169. doi: 10.1247/csf.18035. Epub 2019 Mar 21. Cell Struct Funct. 2019. PMID: 30905922 Review.
Cited by
-
Shining a light on the dark world of plant root-microbe interactions.Proc Natl Acad Sci U S A. 2017 Apr 25;114(17):4281-4283. doi: 10.1073/pnas.1703800114. Epub 2017 Apr 4. Proc Natl Acad Sci U S A. 2017. PMID: 28377510 Free PMC article. No abstract available.
-
In vivo biochemistry: applications for small molecule biosensors in plant biology.Curr Opin Plant Biol. 2013 Jun;16(3):389-95. doi: 10.1016/j.pbi.2013.02.010. Epub 2013 Apr 12. Curr Opin Plant Biol. 2013. PMID: 23587939 Free PMC article. Review.
-
Benzoate mediates the simultaneous repression of anaerobic 4-methylbenzoate and succinate utilization in Magnetospirillum sp. strain pMbN1.BMC Microbiol. 2014 Oct 27;14:269. doi: 10.1186/s12866-014-0269-4. BMC Microbiol. 2014. PMID: 25344702 Free PMC article.
-
Characterization of the l-arabinofuranose-specific GafABCD ABC transporter essential for l-arabinose-dependent growth of the lignocellulose-degrading bacterium Shewanella sp. ANA-3.Microbiology (Reading). 2023 Mar;169(3):001308. doi: 10.1099/mic.0.001308. Microbiology (Reading). 2023. PMID: 36920280 Free PMC article.
-
Optical Sensing Technologies to Elucidate the Interplay between Plant and Microbes.Micromachines (Basel). 2023 Jan 12;14(1):195. doi: 10.3390/mi14010195. Micromachines (Basel). 2023. PMID: 36677256 Free PMC article. Review.
References
-
- Prell J, Poole P (2006) Metabolic changes of rhizobia in legume nodules. Trends Microbiol 14: 161–168. - PubMed
-
- East AK, Mauchline TH, Poole PS (2008) Biosensors for ligand detection. Adv Appl Microbiol 64: 137–166. - PubMed
-
- Davidson AL, Chen J (2004) ATP-Binding cassette transporters in bacteria. Annu Rev Biochem 73: 241–268. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

