Adaptive evolution of the FADS gene cluster within Africa
- PMID: 23028684
- PMCID: PMC3446990
- DOI: 10.1371/journal.pone.0044926
Adaptive evolution of the FADS gene cluster within Africa
Abstract
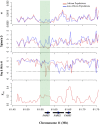
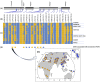
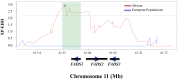
Long chain polyunsaturated fatty acids (LC-PUFAs) are essential for brain structure, development, and function, and adequate dietary quantities of LC-PUFAs are thought to have been necessary for both brain expansion and the increase in brain complexity observed during modern human evolution. Previous studies conducted in largely European populations suggest that humans have limited capacity to synthesize brain LC-PUFAs such as docosahexaenoic acid (DHA) from plant-based medium chain (MC) PUFAs due to limited desaturase activity. Population-based differences in LC-PUFA levels and their product-to-substrate ratios can, in part, be explained by polymorphisms in the fatty acid desaturase (FADS) gene cluster, which have been associated with increased conversion of MC-PUFAs to LC-PUFAs. Here, we show evidence that these high efficiency converter alleles in the FADS gene cluster were likely driven to near fixation in African populations by positive selection ∼85 kya. We hypothesize that selection at FADS variants, which increase LC-PUFA synthesis from plant-based MC-PUFAs, played an important role in allowing African populations obligatorily tethered to marine sources for LC-PUFAs in isolated geographic regions, to rapidly expand throughout the African continent 60-80 kya.
Conflict of interest statement
Figures

References
-
- Macaulay V, Hill C, Achilli A, Rengo C, Clarke D, et al. (2005) Single, rapid coastal settlement of Asia revealed by analysis of complete mitochondrial genomes. Science 308: 1034–1036. - PubMed
-
- Forster P, Matsumura S (2005) Evolution. Did early humans go north or south? Science 308: 965–966. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

