Comprehensive exploration of the effects of miRNA SNPs on monocyte gene expression
- PMID: 23029284
- PMCID: PMC3448685
- DOI: 10.1371/journal.pone.0045863
Comprehensive exploration of the effects of miRNA SNPs on monocyte gene expression
Erratum in
- PLoS One. 2012;7(10). doi:10.1371/annotation/bd5c0312-c902-4065-be2b-385fcf70c125
Abstract
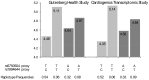
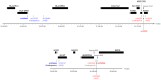
We aimed to assess whether pri-miRNA SNPs (miSNPs) could influence monocyte gene expression, either through marginal association or by interacting with polymorphisms located in 3'UTR regions (3utrSNPs). We then conducted a genome-wide search for marginal miSNPs effects and pairwise miSNPs × 3utrSNPs interactions in a sample of 1,467 individuals for which genome-wide monocyte expression and genotype data were available. Statistical associations that survived multiple testing correction were tested for replication in an independent sample of 758 individuals with both monocyte gene expression and genotype data. In both studies, the hsa-mir-1279 rs1463335 was found to modulate in cis the expression of LYZ and in trans the expression of CNTN6, CTRC, COPZ2, KRT9, LRRFIP1, NOD1, PCDHA6, ST5 and TRAF3IP2 genes, supporting the role of hsa-mir-1279 as a regulator of several genes in monocytes. In addition, we identified two robust miSNPs × 3utrSNPs interactions, one involving HLA-DPB1 rs1042448 and hsa-mir-219-1 rs107822, the second the H1F0 rs1894644 and hsa-mir-659 rs5750504, modulating the expression of the associated genes.As some of the aforementioned genes have previously been reported to reside at disease-associated loci, our findings provide novel arguments supporting the hypothesis that the genetic variability of miRNAs could also contribute to the susceptibility to human diseases.
Conflict of interest statement
Figures

References
-
- Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120: 15–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

