Implication of DNA demethylation and bivalent histone modification for selective gene regulation in mouse primordial germ cells
- PMID: 23029374
- PMCID: PMC3461056
- DOI: 10.1371/journal.pone.0046036
Implication of DNA demethylation and bivalent histone modification for selective gene regulation in mouse primordial germ cells
Abstract
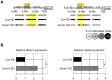
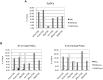
Primordial germ cells (PGCs) sequentially induce specific genes required for their development. We focused on epigenetic changes that regulate PGC-specific gene expression. mil-1, Blimp1, and Stella are preferentially expressed in PGCs, and their expression is upregulated during PGC differentiation. Here, we first determined DNA methylation status of mil-1, Blimp1, and Stella regulatory regions in epiblast and in PGCs, and found that they were hypomethylated in differentiating PGCs after E9.0, in which those genes were highly expressed. We used siRNA to inhibit a maintenance DNA methyltransferase, Dnmt1, in embryonic stem (ES) cells and found that the flanking regions of all three genes became hypomethylated and that expression of each gene increased 1.5- to 3-fold. In addition, we also found 1.5- to 5-fold increase of the PGC genes in the PGCLCs (PGC-like cells) induced form ES cells by knockdown of Dnmt1. We also obtained evidence showing that methylation of the regulatory region of mil-1 resulted in 2.5-fold decrease in expression in a reporter assay. Together, these results suggested that DNA demethylation does not play a major role on initial activation of the PGC genes in the nascent PGCs but contributed to enhancement of their expression in PGCs after E9.0. However, we also found that repression of representative somatic genes, Hoxa1 and Hoxb1, and a tissue-specific gene, Gfap, in PGCs was not dependent on DNA methylation; their flanking regions were hypomethylated, but their expression was not observed in PGCs at E13.5. Their promoter regions showed the bivalent histone modification in PGCs, that may be involved in repression of their expression. Our results indicated that epigenetic status of PGC genes and of somatic genes in PGCs were distinct, and suggested contribution of epigenetic mechanisms in regulation of the expression of a specific gene set in PGCs.
Conflict of interest statement
Figures






References
-
- Ohinata Y, Payer B, O’Carroll D, Ancelin K, Ono Y, et al. (2005) Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436: 207–213. - PubMed
-
- Yamaji M, Seki Y, Kurimoto K, Yabuta Y, Yuasa M, et al. (2008) Critical function of Prdm14 for the establishment of the germ cell lineage in mice. Nat Genet 40: 1016–1022. - PubMed
-
- Yabuta Y, Kurimoto K, Ohinata Y, Seki Y, Saitou M (2006) Gene expression dynamics during germline specification in mice identified by quantitative single-cell gene expression profiling. Biol Reprod 75: 705–716. - PubMed
-
- Okamura D, Tokitake Y, Niwa H, Matsui Y (2008) Requirement of Oct3/4 function for germ cell specification. Dev Biol 317: 576–584. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

