Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development
- PMID: 23031452
- PMCID: PMC3477022
- DOI: 10.1186/1471-2164-13-518
Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development
Abstract
ABBACKGROUND: Early stages of fruit development from initial set through exponential growth are critical determinants of size and yield, however, there has been little detailed analysis of this phase of development. In this study we combined morphological analysis with 454 pyrosequencing to study transcript level changes occurring in young cucumber fruit at five ages from anthesis through the end of exponential growth.
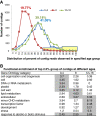
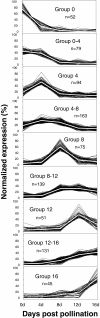
Results: The fruit samples produced 1.13 million ESTs which were assembled into 27,859 contigs with a mean length of 834 base pairs and a mean of 67 reads per contig. All contigs were mapped to the cucumber genome. Principal component analysis separated the fruit ages into three groups corresponding with cell division/pre-exponential growth (0 and 4 days post pollination (dpp)), peak exponential expansion (8dpp), and late/post-exponential expansion stages of growth (12 and 16 dpp). Transcripts predominantly expressed at 0 and 4 dpp included homologs of histones, cyclins, and plastid and photosynthesis related genes. The group of genes with peak transcript levels at 8dpp included cytoskeleton, cell wall, lipid metabolism and phloem related proteins. This group was also dominated by genes with unknown function or without known homologs outside of cucurbits. A second shift in transcript profile was observed at 12-16dpp, which was characterized by abiotic and biotic stress related genes and significant enrichment for transcription factor gene homologs, including many associated with stress response and development.
Conclusions: The transcriptome data coupled with morphological analyses provide an informative picture of early fruit development. Progressive waves of transcript abundance were associated with cell division, development of photosynthetic capacity, cell expansion and fruit growth, phloem activity, protection of the fruit surface, and finally transition away from fruit growth toward a stage of enhanced stress responses. These results suggest that the interval between expansive growth and ripening includes further developmental differentiation with an emphasis on defense. The increased transcript levels of cucurbit-specific genes during the exponential growth stage may indicate unique factors contributing to rapid growth in cucurbits.
Figures





References
-
- Boonkorkaew P, Hikosaka S, Sugiyama N. Effect of pollination on cell division, cell enlargement, and endogenous hormones in fruit development in a gynoecious cucumber. Scientia Hortic. 2008;116:1–7. doi: 10.1016/j.scienta.2007.10.027. - DOI
-
- Marcelis LFM, Hofman-Eijer LRB. Cell division and expansion in the cucumber fruit. J Hortic Sci. 1993;68:665–671.
-
- Robinson RW, Decker-Walters DS. Cucurbits. CAB International, New York, NY; 1997.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

