Role of protons in sugar binding to LacY
- PMID: 23033496
- PMCID: PMC3479522
- DOI: 10.1073/pnas.1214890109
Role of protons in sugar binding to LacY
Abstract
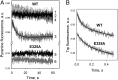
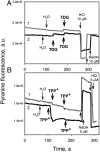
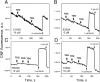
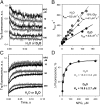
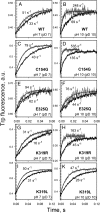
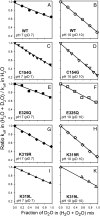
WT lactose permease of Escherichia coli (LacY) reconstituted into proteoliposomes loaded with a pH-sensitive fluorophore exhibits robust uphill H(+) translocation coupled with downhill lactose transport. However, galactoside binding by mutants defective in lactose-induced H(+) translocation is not accompanied by release of an H(+) on the interior of the proteoliposomes. Because the pK(a) value for galactoside binding is ∼10.5, protonation of LacY likely precedes sugar binding at physiological pH. Consistently, purified WT LacY, as well as the mutants, binds substrate at pH 7.5-8.5 in detergent, but no change in ambient pH is observed, demonstrating directly that LacY already is protonated when sugar binds. However, a kinetic isotope effect (KIE) on the rate of binding is observed, indicating that deuterium substitution for protium affects an H(+) transfer reaction within LacY that is associated with sugar binding. At neutral pH or pD, both the rate of sugar dissociation (k(off)) and the forward rate (k(on)) are slower in D(2)O than in H(2)O (KIE is ∼2), and, as a result, no change in affinity (K(d)) is observed. Alkaline conditions enhance the effect of D(2)O on k(off), the KIE increases to 3.6-4.0, and affinity for sugar increases compared with H(2)O. In contrast, LacY mutants that exhibit pH-independent high-affinity binding up to pH 11.0 (e.g., Glu325 → Gln) exhibit the same KIE (1.5-1.8) at neutral or alkaline pH (pD). Proton inventory studies exhibit a linear relationship between k(off) and D(2)O concentration at neutral and alkaline pH, indicating that internal transfer of a single H(+) is involved in the KIE.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Abramson J, et al. Structure and mechanism of the lactose permease of Escherichia coli. Science. 2003;301:610–615. - PubMed
-
- Frillingos S, Sahin-Tóth M, Wu J, Kaback HR. Cys-scanning mutagenesis: A novel approach to structure function relationships in polytopic membrane proteins. FASEB J. 1998;12:1281–1299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

