Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia
- PMID: 23034282
- PMCID: PMC3512237
- DOI: 10.1182/blood-2012-05-429506
Selective inhibitors of nuclear export show that CRM1/XPO1 is a target in chronic lymphocytic leukemia
Abstract
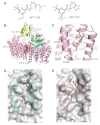
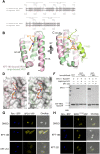
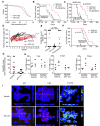
The nuclear export protein XPO1 is overexpressed in cancer, leading to the cytoplasmic mislocalization of multiple tumor suppressor proteins. Existing XPO1-targeting agents lack selectivity and have been associated with significant toxicity. Small molecule selective inhibitors of nuclear export (SINEs) were designed that specifically inhibit XPO1. Genetic experiments and X-ray structures demonstrate that SINE covalently bind to a cysteine residue in the cargo-binding groove of XPO1, thereby inhibiting nuclear export of cargo proteins. The clinical relevance of SINEs was explored in chronic lymphocytic leukemia (CLL), a disease associated with recurrent XPO1 mutations. Evidence is presented that SINEs can restore normal regulation to the majority of the dysregulated pathways in CLL both in vitro and in vivo and induce apoptosis of CLL cells with a favorable therapeutic index, with enhanced killing of genomically high-risk CLL cells that are typically unresponsive to traditional therapies. More importantly, SINE slows disease progression, and improves overall survival in the Eμ-TCL1-SCID mouse model of CLL with minimal weight loss or other toxicities. Together, these findings demonstrate that XPO1 is a valid target in CLL with minimal effects on normal cells and provide a basis for the development of SINEs in CLL and related hematologic malignancies.
Figures




References
-
- Yao Y, Dong Y, Lin F, et al. The expression of CRM1 is associated with prognosis in human osteosarcoma. Oncol Rep. 2009;21(1):229–235. - PubMed
-
- Monecke T, Guttler T, Neumann P, Dickmanns A, Gorlich D, Ficner R. Crystal structure of the nuclear export receptor CRM1 in complex with Snurportin1 and RanGTP. Science. 2009;324(5930):1087–1091. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

