Ribozyme-based insulator parts buffer synthetic circuits from genetic context
- PMID: 23034349
- PMCID: PMC3914141
- DOI: 10.1038/nbt.2401
Ribozyme-based insulator parts buffer synthetic circuits from genetic context
Abstract
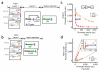
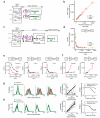
Synthetic genetic programs are built from circuits that integrate sensors and implement temporal control of gene expression. Transcriptional circuits are layered by using promoters to carry the signal between circuits. In other words, the output promoter of one circuit serves as the input promoter to the next. Thus, connecting circuits requires physically connecting a promoter to the next circuit. We show that the sequence at the junction between the input promoter and circuit can affect the input-output response (transfer function) of the circuit. A library of putative sequences that might reduce (or buffer) such context effects, which we refer to as 'insulator parts', is screened in Escherichia coli. We find that ribozymes that cleave the 5' untranslated region (5'-UTR) of the mRNA are effective insulators. They generate quantitatively identical transfer functions, irrespective of the identity of the input promoter. When these insulators are used to join synthetic gene circuits, the behavior of layered circuits can be predicted using a mathematical model. The inclusion of insulators will be critical in reliably permuting circuits to build different programs.
Figures

Comment in
-
Insulating gene circuits from context by RNA processing.Nat Biotechnol. 2012 Nov;30(11):1061-2. doi: 10.1038/nbt.2411. Nat Biotechnol. 2012. PMID: 23138300 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

