Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder
- PMID: 23034409
- PMCID: PMC3530681
- DOI: 10.1101/gr.141887.112
Small noncoding differentially methylated copy-number variants, including lncRNA genes, cause a lethal lung developmental disorder
Abstract
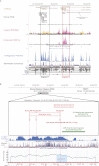
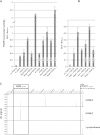
An unanticipated and tremendous amount of the noncoding sequence of the human genome is transcribed. Long noncoding RNAs (lncRNAs) constitute a significant fraction of non-protein-coding transcripts; however, their functions remain enigmatic. We demonstrate that deletions of a small noncoding differentially methylated region at 16q24.1, including lncRNA genes, cause a lethal lung developmental disorder, alveolar capillary dysplasia with misalignment of pulmonary veins (ACD/MPV), with parent-of-origin effects. We identify overlapping deletions 250 kb upstream of FOXF1 in nine patients with ACD/MPV that arose de novo specifically on the maternally inherited chromosome and delete lung-specific lncRNA genes. These deletions define a distant cis-regulatory region that harbors, besides lncRNA genes, also a differentially methylated CpG island, binds GLI2 depending on the methylation status of this CpG island, and physically interacts with and up-regulates the FOXF1 promoter. We suggest that lung-transcribed 16q24.1 lncRNAs may contribute to long-range regulation of FOXF1 by GLI2 and other transcription factors. Perturbation of lncRNA-mediated chromatin interactions may, in general, be responsible for position effect phenomena and potentially cause many disorders of human development.
Figures


References
-
- Chang VW, Ho Y 2001. Structural characterization of the mouse Foxf1a gene. Gene 267: 201–211 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
