Birth and expression evolution of mammalian microRNA genes
- PMID: 23034410
- PMCID: PMC3530682
- DOI: 10.1101/gr.140269.112
Birth and expression evolution of mammalian microRNA genes
Abstract
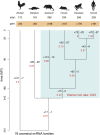
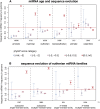
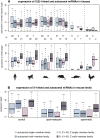
MicroRNAs (miRNAs) are major post-transcriptional regulators of gene expression, yet their origins and functional evolution in mammals remain little understood due to the lack of appropriate comparative data. Using RNA sequencing, we have generated extensive and comparable miRNA data for five organs in six species that represent all main mammalian lineages and birds (the evolutionary outgroup) with the aim to unravel the evolution of mammalian miRNAs. Our analyses reveal an overall expansion of miRNA repertoires in mammals, with threefold accelerated birth rates of miRNA families in placentals and marsupials, facilitated by the de novo emergence of miRNAs in host gene introns. Generally, our analyses suggest a high rate of miRNA family turnover in mammals with many newly emerged miRNA families being lost soon after their formation. Selectively preserved mammalian miRNA families gradually evolved higher expression levels, as well as altered mature sequences and target gene repertoires, and were apparently mainly recruited to exert regulatory functions in nervous tissues. However, miRNAs that originated on the X chromosome evolved high expression levels and potentially diverse functions during spermatogenesis, including meiosis, through selectively driven duplication-divergence processes. Overall, our study thus provides detailed insights into the birth and evolution of mammalian miRNA genes and the associated selective forces.
Figures


References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ 1990. Basic local alignment search tool. J Mol Biol 215: 403–410 - PubMed
-
- Benjamini Y, Hochberg Y 1995. Controlling the false discovery rate: A practical and powerful approach to multiple testing. JR Stat Soc 57: 289–300
-
- Berezikov E 2011. Evolution of microRNA diversity and regulation in animals. Nat Rev Genet 12: 846–860 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
