TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function
- PMID: 23035106
- PMCID: PMC3978779
- DOI: 10.1523/JNEUROSCI.2433-12.2012
TNiK is required for postsynaptic and nuclear signaling pathways and cognitive function
Abstract
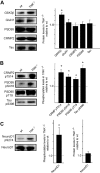
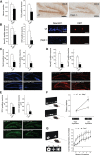
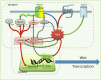
Traf2 and NcK interacting kinase (TNiK) contains serine-threonine kinase and scaffold domains and has been implicated in cell proliferation and glutamate receptor regulation in vitro. Here we report its role in vivo using mice carrying a knock-out mutation. TNiK binds protein complexes in the synapse linking it to the NMDA receptor (NMDAR) via AKAP9. NMDAR and metabotropic receptors bidirectionally regulate TNiK phosphorylation and TNiK is required for AMPA expression and synaptic function. TNiK also organizes nuclear complexes and in the absence of TNiK, there was a marked elevation in GSK3β and phosphorylation levels of its cognate phosphorylation sites on NeuroD1 with alterations in Wnt pathway signaling. We observed impairments in dentate gyrus neurogenesis in TNiK knock-out mice and cognitive testing using the touchscreen apparatus revealed impairments in pattern separation on a test of spatial discrimination. Object-location paired associate learning, which is dependent on glutamatergic signaling, was also impaired. Additionally, TNiK knock-out mice displayed hyperlocomotor behavior that could be rapidly reversed by GSK3β inhibitors, indicating the potential for pharmacological rescue of a behavioral phenotype. These data establish TNiK as a critical regulator of cognitive functions and suggest it may play a regulatory role in diseases impacting on its interacting proteins and complexes.
Figures





References
-
- Ayalew M, Le-Niculescu H, Levey DF, Jain N, Changala B, Patel SD, Winiger E, Breier A, Shekhar A, Amdur R, Koller D, Nurnberger JI, Corvin A, Geyer M, Tsuang MT, Salomon D, Schork NJ, Fanous AH, O'Donovan MC, Niculescu AB. Convergent functional genomics of schizophrenia: from comprehensive understanding to genetic risk prediction. Mol Psychiatry. 2012;17:887–905. - PMC - PubMed
