A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function
- PMID: 23035249
- PMCID: PMC3479514
- DOI: 10.1073/pnas.1209751109
A fundamental protein property, thermodynamic stability, revealed solely from large-scale measurements of protein function
Abstract
The ability of a protein to carry out a given function results from fundamental physicochemical properties that include the protein's structure, mechanism of action, and thermodynamic stability. Traditional approaches to study these properties have typically required the direct measurement of the property of interest, oftentimes a laborious undertaking. Although protein properties can be probed by mutagenesis, this approach has been limited by its low throughput. Recent technological developments have enabled the rapid quantification of a protein's function, such as binding to a ligand, for numerous variants of that protein. Here, we measure the ability of 47,000 variants of a WW domain to bind to a peptide ligand and use these functional measurements to identify stabilizing mutations without directly assaying stability. Our approach is rooted in the well-established concept that protein function is closely related to stability. Protein function is generally reduced by destabilizing mutations, but this decrease can be rescued by stabilizing mutations. Based on this observation, we introduce partner potentiation, a metric that uses this rescue ability to identify stabilizing mutations, and identify 15 candidate stabilizing mutations in the WW domain. We tested six candidates by thermal denaturation and found two highly stabilizing mutations, one more stabilizing than any previously known mutation. Thus, physicochemical properties such as stability are latent within these large-scale protein functional data and can be revealed by systematic analysis. This approach should allow other protein properties to be discovered.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
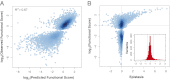

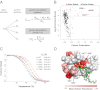
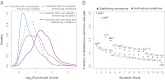
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

