DMET-analyzer: automatic analysis of Affymetrix DMET data
- PMID: 23035929
- PMCID: PMC3496574
- DOI: 10.1186/1471-2105-13-258
DMET-analyzer: automatic analysis of Affymetrix DMET data
Abstract
Background: Clinical Bioinformatics is currently growing and is based on the integration of clinical and omics data aiming at the development of personalized medicine. Thus the introduction of novel technologies able to investigate the relationship among clinical states and biological machineries may help the development of this field. For instance the Affymetrix DMET platform (drug metabolism enzymes and transporters) is able to study the relationship among the variation of the genome of patients and drug metabolism, detecting SNPs (Single Nucleotide Polymorphism) on genes related to drug metabolism. This may allow for instance to find genetic variants in patients which present different drug responses, in pharmacogenomics and clinical studies. Despite this, there is currently a lack in the development of open-source algorithms and tools for the analysis of DMET data. Existing software tools for DMET data generally allow only the preprocessing of binary data (e.g. the DMET-Console provided by Affymetrix) and simple data analysis operations, but do not allow to test the association of the presence of SNPs with the response to drugs.
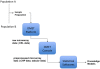
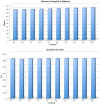
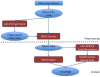
Results: We developed DMET-Analyzer a tool for the automatic association analysis among the variation of the patient genomes and the clinical conditions of patients, i.e. the different response to drugs. The proposed system allows: (i) to automatize the workflow of analysis of DMET-SNP data avoiding the use of multiple tools; (ii) the automatic annotation of DMET-SNP data and the search in existing databases of SNPs (e.g. dbSNP), (iii) the association of SNP with pathway through the search in PharmaGKB, a major knowledge base for pharmacogenomic studies. DMET-Analyzer has a simple graphical user interface that allows users (doctors/biologists) to upload and analyse DMET files produced by Affymetrix DMET-Console in an interactive way. The effectiveness and easy use of DMET Analyzer is demonstrated through different case studies regarding the analysis of clinical datasets produced in the University Hospital of Catanzaro, Italy.
Conclusion: DMET Analyzer is a novel tool able to automatically analyse data produced by the DMET-platform in case-control association studies. Using such tool user may avoid wasting time in the manual execution of multiple statistical tests avoiding possible errors and reducing the amount of time needed for a whole experiment. Moreover annotations and the direct link to external databases may increase the biological knowledge extracted. The system is freely available for academic purposes at: https://sourceforge.net/projects/dmetanalyzer/files/
Figures

References
-
- Knaup P, Ammenwerth E, Brandner R, Brigl B, Fischer G, Garde S, Lang E, Pilgram R, Ruderich F, Singer R, Wolff A, Haux R, Kulikowski C. Towards clinical bioinformatics: advancing genomic medicine with informatics methods and tools. Methods Inf Med. 2004;43:302–307. - PubMed
-
- Martin-Sanchez F, Iakovidis I, Norager S, Maojo V, de Groen P, Van der L, Jones T, Abraham-Fuchs K, Apweiler R, Babic A, Baud R, Breton V, Cinquin P, Doupi P, Dugas M, Eils R, Engelbrecht R, Ghazal P, Jehenson P, Kulikowski C, Lampe K, De Moor G, Orphanoudakis S, Rossing N, Sarachan B, Sousa A, Spekowius G, Thireos G, Zahlmann G, Zvarova J. et al.Synergy between medical informatics and bioinformatics: facilitating genomic medicine for future health care. J Biomed Inform. 2004;37:30–42. doi: 10.1016/j.jbi.2003.09.003. - DOI - PubMed
-
- Wang X, Liotta L. Clinical bioinformatics: a new emerging science. J Clin Bioinformatics. 2011;1:1. doi: 10.1186/2043-9113-1-1. [ http://www.jclinbioinformatics.com/content/1/1/1] - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

