Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair
- PMID: 23038248
- PMCID: PMC3501068
- DOI: 10.1074/jbc.M112.393504
Poly(ADP-ribose) contributes to an association between poly(ADP-ribose) polymerase-1 and xeroderma pigmentosum complementation group A in nucleotide excision repair
Abstract
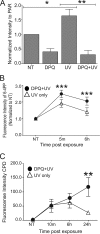
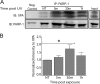
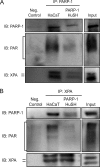
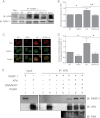
Exposure to ultraviolet radiation (UVR) promotes the formation of UVR-induced, DNA helix distorting photolesions such as (6-4) pyrimidine-pyrimidone photoproducts and cyclobutane pyrimidine dimers. Effective repair of such lesions by the nucleotide excision repair (NER) pathway is required to prevent DNA mutations and chromosome aberrations. Poly(ADP-ribose) polymerase-1 (PARP-1) is a zinc finger protein with well documented involvement in base excision repair. PARP-1 is activated in response to DNA damage and catalyzes the formation of poly(ADP-ribose) subunits that assist in the assembly of DNA repair proteins at sites of damage. In this study, we present evidence for PARP-1 contributions to NER, extending the knowledge of PARP-1 function in DNA repair beyond the established role in base excision repair. Silencing the PARP-1 protein or inhibiting PARP activity leads to retention of UVR-induced photolesions. PARP activation following UVR exposure promotes association between PARP-1 and XPA, a central protein in NER. Administration of PARP inhibitors confirms that poly(ADP-ribose) facilitates PARP-1 association with XPA in whole cell extracts, in isolated chromatin complexes, and in vitro. Furthermore, inhibition of PARP activity decreases UVR-stimulated XPA chromatin association, illustrating that these relationships occur in a meaningful context for NER. These results provide a mechanistic link for PARP activity in the repair of UVR-induced photoproducts.
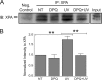
Figures


Similar articles
-
Differential sensitivities of cellular XPA and PARP-1 to arsenite inhibition and zinc rescue.Toxicol Appl Pharmacol. 2017 Sep 15;331:108-115. doi: 10.1016/j.taap.2017.05.031. Epub 2017 May 25. Toxicol Appl Pharmacol. 2017. PMID: 28552776 Free PMC article.
-
Inhibition of poly(ADP-ribose) polymerase-1 by arsenite interferes with repair of oxidative DNA damage.J Biol Chem. 2009 Mar 13;284(11):6809-17. doi: 10.1074/jbc.M805566200. Epub 2008 Dec 3. J Biol Chem. 2009. PMID: 19056730 Free PMC article.
-
Poly(ADP-ribose)-mediated interplay of XPA and PARP1 leads to reciprocal regulation of protein function.FEBS J. 2014 Aug;281(16):3625-41. doi: 10.1111/febs.12885. Epub 2014 Jul 21. FEBS J. 2014. PMID: 24953096 Free PMC article.
-
Modulating poly (ADP-ribose) polymerase activity: potential for the prevention and therapy of pathogenic situations involving DNA damage and oxidative stress.Curr Pharm Biotechnol. 2002 Sep;3(3):275-83. doi: 10.2174/1389201023378265. Curr Pharm Biotechnol. 2002. PMID: 12164482 Review.
-
Structure and function of poly(ADP-ribose) polymerase-1: role in oxidative stress-related pathologies.Curr Vasc Pharmacol. 2005 Jul;3(3):209-14. doi: 10.2174/1570161054368625. Curr Vasc Pharmacol. 2005. PMID: 16026317 Review.
Cited by
-
Regulation of therapeutic resistance in cancers by receptor tyrosine kinases.Am J Cancer Res. 2016 Mar 15;6(4):827-42. eCollection 2016. Am J Cancer Res. 2016. PMID: 27186434 Free PMC article.
-
H4K20me2: Orchestrating the recruitment of DNA repair factors in nucleotide excision repair.Nucleus. 2018 Jan 1;9(1):212-215. doi: 10.1080/19491034.2018.1444327. Nucleus. 2018. PMID: 29482435 Free PMC article.
-
Contribution of NADPH oxidase to the retention of UVR-induced DNA damage by arsenic.Toxicol Appl Pharmacol. 2022 Jan 1;434:115799. doi: 10.1016/j.taap.2021.115799. Epub 2021 Nov 16. Toxicol Appl Pharmacol. 2022. PMID: 34798142 Free PMC article.
-
Melanocytotoxic chemicals and their toxic mechanisms.Toxicol Res. 2022 Aug 22;38(4):417-435. doi: 10.1007/s43188-022-00144-2. eCollection 2022 Oct. Toxicol Res. 2022. PMID: 36277364 Free PMC article. Review.
-
Melanocytes and keratinocytes have distinct and shared responses to ultraviolet radiation and arsenic.Toxicol Lett. 2014 Jan 30;224(3):407-15. doi: 10.1016/j.toxlet.2013.11.010. Epub 2013 Nov 21. Toxicol Lett. 2014. PMID: 24270004 Free PMC article.
References
-
- Narayanan D. L., Saladi R. N., Fox J. L. (2010) Ultraviolet radiation and skin cancer. Int. J. Dermatol. 49, 978–986 - PubMed
-
- Park C. J., Choi B. S. (2006) The protein shuffle. Sequential interactions among components of the human nucleotide excision repair pathway. FEBS J. 273, 1600–1608 - PubMed
-
- Pfeifer G. P., You Y. H., Besaratinia A. (2005) Mutations induced by ultraviolet light. Mutat. Res. 571, 19–31 - PubMed
-
- Sinha R. P., Häder D. P. (2002) UV-induced DNA damage and repair. A review. Photochem. Photobiol. Sci. 1, 225–236 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

