Mitochondrial SKN-1/Nrf mediates a conserved starvation response
- PMID: 23040073
- PMCID: PMC3774140
- DOI: 10.1016/j.cmet.2012.09.007
Mitochondrial SKN-1/Nrf mediates a conserved starvation response
Abstract
SKN-1/Nrf plays multiple essential roles in development and cellular homeostasis. We demonstrate that SKN-1 executes a specific and appropriate transcriptional response to changes in available nutrients, leading to metabolic adaptation. We isolated gain-of-function (gf) alleles of skn-1, affecting a domain of SKN-1 that binds the transcription factor MXL-3 and the mitochondrial outer membrane protein PGAM-5. These skn-1(gf) mutants perceive a state of starvation even in the presence of plentiful food. The aberrant monitoring of cellular nutritional status leads to an altered survival response in which skn-1(gf) mutants transcriptionally activate genes associated with metabolism, adaptation to starvation, aging, and survival. The triggered starvation response is conserved in mice with constitutively activated Nrf and may contribute to the tumorgenicity associated with activating Nrf mutations in mammalian somatic cells. Our findings delineate an evolutionarily conserved metabolic axis of SKN-1/Nrf, further establishing the complexity of this pathway.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
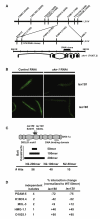
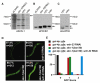
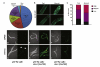

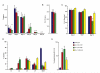
References
-
- Bauer M, Hamm AC, Bonaus M, Jacob A, Jaekel J, Schorle H, Pankratz MJ, Katzenberger JD. Starvation response in mouse liver shows strong correlation with life-span-prolonging processes. Physiol. Genomics. 2004;17:230–244. - PubMed
-
- Baugh LR, Demodena J, Sternberg PW. RNA Pol II accumulates at promoters of growth genes during developmental arrest. Science. 2009;324:92–94. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

