The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses
- PMID: 23041064
- PMCID: PMC3717271
- DOI: 10.1016/j.immuni.2012.09.007
The transcription factor T-bet is induced by multiple pathways and prevents an endogenous Th2 cell program during Th1 cell responses
Abstract
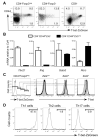
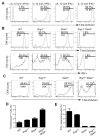
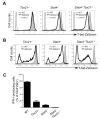
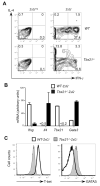
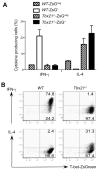
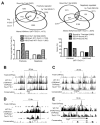
T-bet is a critical transcription factor for T helper 1 (Th1) cell differentiation. To study the regulation and functions of T-bet, we developed a T-bet-ZsGreen reporter mouse strain. We determined that interleukin-12 (IL-12) and interferon-γ (IFN-γ) were redundant in inducing T-bet in mice infected with Toxoplasma gondii and that T-bet did not contribute to its own expression when induced by IL-12 and IFN-γ. By contrast, T-bet and the transcription factor Stat4 were critical for IFN-γ production whereas IFN-γ signaling was dispensable for inducing IFN-γ. Loss of T-bet resulted in activation of an endogenous program driving Th2 cell differentiation in cells expressing T-bet-ZsGreen. Genome-wide analyses indicated that T-bet directly induced many Th1 cell-related genes but indirectly suppressed Th2 cell-related genes. Our study revealed redundancy and synergy among several Th1 cell-inducing pathways in regulating the expression of T-bet and IFN-γ, and a critical role of T-bet in suppressing an endogenous Th2 cell-associated program.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures

Comment in
-
New tricks from an old master.Immunity. 2012 Oct 19;37(4):591-3. doi: 10.1016/j.immuni.2012.10.004. Immunity. 2012. PMID: 23084353 Free PMC article.
References
-
- Afkarian M, Sedy JR, Yang J, Jacobson NG, Cereb N, Yang SY, Murphy TL, Murphy KM. T-bet is a STAT1-induced regulator of IL-12R expression in naive CD4+ T cells. Nat. Immunol. 2002;3:549–557. - PubMed
-
- Ansel KM, Djuretic I, Tanasa B, Rao A. Regulation of Th2 differentiation and Il4 locus accessibility. Annu. Rev. Immunol. 2006;24:607–656. - PubMed
-
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. - PubMed
-
- Finkelman FD, Morris SC, Orekhova T, Mori M, Donaldson D, Reiner SL, Reilly NL, Schopf L, Urban JF., Jr. Stat6 regulation of in vivo IL-4 responses. J. Immunol. 2000;164:2303–2310. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

