Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies
- PMID: 23041239
- PMCID: PMC3490334
- DOI: 10.1016/S1474-4422(12)70234-X
Genetic risk factors for ischaemic stroke and its subtypes (the METASTROKE collaboration): a meta-analysis of genome-wide association studies
Erratum in
-
Corrections.Lancet Neurol. 2015 Aug;14(8):788. doi: 10.1016/S1474-4422(15)00155-6. Lancet Neurol. 2015. PMID: 26194924 Free PMC article. No abstract available.
Abstract
Background: Various genome-wide association studies (GWAS) have been done in ischaemic stroke, identifying a few loci associated with the disease, but sample sizes have been 3500 cases or less. We established the METASTROKE collaboration with the aim of validating associations from previous GWAS and identifying novel genetic associations through meta-analysis of GWAS datasets for ischaemic stroke and its subtypes.
Methods: We meta-analysed data from 15 ischaemic stroke cohorts with a total of 12 389 individuals with ischaemic stroke and 62 004 controls, all of European ancestry. For the associations reaching genome-wide significance in METASTROKE, we did a further analysis, conditioning on the lead single nucleotide polymorphism in every associated region. Replication of novel suggestive signals was done in 13 347 cases and 29 083 controls.
Findings: We verified previous associations for cardioembolic stroke near PITX2 (p=2·8×10(-16)) and ZFHX3 (p=2·28×10(-8)), and for large-vessel stroke at a 9p21 locus (p=3·32×10(-5)) and HDAC9 (p=2·03×10(-12)). Additionally, we verified that all associations were subtype specific. Conditional analysis in the three regions for which the associations reached genome-wide significance (PITX2, ZFHX3, and HDAC9) indicated that all the signal in each region could be attributed to one risk haplotype. We also identified 12 potentially novel loci at p<5×10(-6). However, we were unable to replicate any of these novel associations in the replication cohort.
Interpretation: Our results show that, although genetic variants can be detected in patients with ischaemic stroke when compared with controls, all associations we were able to confirm are specific to a stroke subtype. This finding has two implications. First, to maximise success of genetic studies in ischaemic stroke, detailed stroke subtyping is required. Second, different genetic pathophysiological mechanisms seem to be associated with different stroke subtypes.
Funding: Wellcome Trust, UK Medical Research Council (MRC), Australian National and Medical Health Research Council, National Institutes of Health (NIH) including National Heart, Lung and Blood Institute (NHLBI), the National Institute on Aging (NIA), the National Human Genome Research Institute (NHGRI), and the National Institute of Neurological Disorders and Stroke (NINDS).
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures
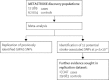

Comment in
-
From genes to stroke subtypes.Lancet Neurol. 2012 Nov;11(11):931-3. doi: 10.1016/S1474-4422(12)70235-1. Epub 2012 Oct 5. Lancet Neurol. 2012. PMID: 23041236 No abstract available.
-
Risk factors and cerebrovascular disease.J Neurol. 2013 Feb;260(2):692-4. doi: 10.1007/s00415-013-6835-0. J Neurol. 2013. PMID: 23355176 No abstract available.
References
-
- Department of Health . Reducing brain damage: faster access to better stroke care. National Audit Office; London: 2005.
-
- Sacco RL, Ellenberg JH, Mohr JP. Infarcts of undetermined cause: the NINCDS Stroke Data Bank. Ann Neurol. 1989;25:382–390. - PubMed
-
- Dichgans M. Genetics of ischaemic stroke. Lancet Neurol. 2007;6:149–161. - PubMed
-
- Dichgans M, Markus HS. Genetic association studies in stroke: methodological issues and proposed standard criteria. Stroke. 2005;36:2027–2031. - PubMed
Publication types
MeSH terms
Grants and funding
- N01 HC055020/HL/NHLBI NIH HHS/United States
- R01 HL080295/HL/NHLBI NIH HHS/United States
- BB/F019394/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- R56 AG023629/AG/NIA NIH HHS/United States
- U54 RR020278/RR/NCRR NIH HHS/United States
- Z01 AG000015/ImNIH/Intramural NIH HHS/United States
- HHSN268201100012C/HL/NHLBI NIH HHS/United States
- U01 HG005160/HG/NHGRI NIH HHS/United States
- HHSN268201100009I/HL/NHLBI NIH HHS/United States
- N01 HC055016/HL/NHLBI NIH HHS/United States
- R01 NS045012/NS/NINDS NIH HHS/United States
- R01 NS017950/NS/NINDS NIH HHS/United States
- R01 NS042733/NS/NINDS NIH HHS/United States
- U01 HG004446/HG/NHGRI NIH HHS/United States
- Z01 AG000954/ImNIH/Intramural NIH HHS/United States
- N01 HC055019/HL/NHLBI NIH HHS/United States
- 095626/WT_/Wellcome Trust/United Kingdom
- HHSN268201100010C/HL/NHLBI NIH HHS/United States
- UL1 RR025005/RR/NCRR NIH HHS/United States
- R01 AG015928/AG/NIA NIH HHS/United States
- HHSN268201100008C/HL/NHLBI NIH HHS/United States
- U01 HL080295/HL/NHLBI NIH HHS/United States
- R01 HL087676/HL/NHLBI NIH HHS/United States
- HHSN268201100005G/HL/NHLBI NIH HHS/United States
- U01 HL096917/HL/NHLBI NIH HHS/United States
- HHSN268201100008I/HL/NHLBI NIH HHS/United States
- R01 HL059367/HL/NHLBI NIH HHS/United States
- N01 HC055021/HL/NHLBI NIH HHS/United States
- HHSN268201100007C/HL/NHLBI NIH HHS/United States
- N01 HC085086/HL/NHLBI NIH HHS/United States
- R01 HL085251/HL/NHLBI NIH HHS/United States
- R56 AG020098/AG/NIA NIH HHS/United States
- HHSN268201100011I/HL/NHLBI NIH HHS/United States
- HHSN268201100011C/HL/NHLBI NIH HHS/United States
- R01 HL086694/HL/NHLBI NIH HHS/United States
- G0500987/MRC_/Medical Research Council/United Kingdom
- N01 HC055015/HL/NHLBI NIH HHS/United States
- R01 HL087652/HL/NHLBI NIH HHS/United States
- U01 HG004402/HG/NHGRI NIH HHS/United States
- U01 HG004424/HG/NHGRI NIH HHS/United States
- UL1 TR000124/TR/NCATS NIH HHS/United States
- MR/K026992/1/MRC_/Medical Research Council/United Kingdom
- MC_U137686857/MRC_/Medical Research Council/United Kingdom
- R01 HL093029/HL/NHLBI NIH HHS/United States
- P 20545/FWF_/Austrian Science Fund FWF/Austria
- R01 HL105756/HL/NHLBI NIH HHS/United States
- G0601325/MRC_/Medical Research Council/United Kingdom
- P30 DK063491/DK/NIDDK NIH HHS/United States
- HHSN268200782096C/HG/NHGRI NIH HHS/United States
- HHSN268201100006C/HL/NHLBI NIH HHS/United States
- R01 AG008122/AG/NIA NIH HHS/United States
- HHSN268201200036C/HL/NHLBI NIH HHS/United States
- P30 DK072488/DK/NIDDK NIH HHS/United States
- N01 HC025195/HL/NHLBI NIH HHS/United States
- R01 AG033193/AG/NIA NIH HHS/United States
- U01 NS069208/NS/NINDS NIH HHS/United States
- R01 NS039987/NS/NINDS NIH HHS/United States
- N01 HC055222/HL/NHLBI NIH HHS/United States
- HHSN268201100005I/HL/NHLBI NIH HHS/United States
- RP-PG-0606-1146/DH_/Department of Health/United Kingdom
- N01 HC085079/HL/NHLBI NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- U01 HG005157/HG/NHGRI NIH HHS/United States
- N01 HC055018/HL/NHLBI NIH HHS/United States
- R01 AG020098/AG/NIA NIH HHS/United States
- R01 NS034447/NS/NINDS NIH HHS/United States
- U01 HG004436/HG/NHGRI NIH HHS/United States
- HHSN268201100009C/HL/NHLBI NIH HHS/United States
- OSRP2/1006/DMT_/The Dunhill Medical Trust/United Kingdom
- HHSN268201100005C/HL/NHLBI NIH HHS/United States
- G0700704/MRC_/Medical Research Council/United Kingdom
- HHSN268201100007I/HL/NHLBI NIH HHS/United States
- N01 HC055022/HL/NHLBI NIH HHS/United States
- N01 HC075150/HL/NHLBI NIH HHS/United States
- R01 AG023629/AG/NIA NIH HHS/United States
- R01 HL087641/HL/NHLBI NIH HHS/United States
- R01 HL073410/HL/NHLBI NIH HHS/United States
- U01 HG005152/HG/NHGRI NIH HHS/United States
- R01 AG027058/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

