Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants
- PMID: 23042297
- PMCID: PMC3615175
- DOI: 10.1002/cne.23242
Loss of Gsx1 and Gsx2 function rescues distinct phenotypes in Dlx1/2 mutants
Erratum in
-
Erratum.J Comp Neurol. 2024 Feb;532(2):e25592. doi: 10.1002/cne.25592. J Comp Neurol. 2024. PMID: 38362770 Free PMC article. No abstract available.
Abstract
Mice lacking the Dlx1 and Dlx2 homeobox genes (Dlx1/2 mutants) have severe deficits in subpallial differentiation, including overexpression of the Gsx1 and Gsx2 homeobox genes. To investigate whether Gsx overexpression contributes to the Dlx1/2 mutant phenotypes, we made compound loss-of-function mutants. Eliminating Gsx2 function from the Dlx1/2 mutants rescued the increased expression of Ascl1 and Hes5 (Notch signaling mediators) and Olig2 (oligodendrogenesis mediator). In addition, Dlx1/2;Gsx2 mutants, like Dlx1/2;Ascl1 mutants, exacerbated the Gsx2 and Dlx1/2 patterning and differentiation phenotypes, particularly in the lateral ganglionic eminence (LGE) caudal ganglionic eminence (CGE), and septum, including loss of GAD1 expression. On the other hand, eliminating Gsx1 function from the Dlx1/2 mutants (Dlx1/2;Gsx1 mutants) did not severely exacerbate their phenotype; on the contrary, it resulted in a partial rescue of medial ganglionic eminence (MGE) properties, including interneuron migration to the cortex. Thus, despite their redundant properties, Gsx1 and -2 have distinct interactions with Dlx1 and -2. Gsx2 interaction is strongest in the LGE, CGE, and septum, whereas the Gsx1 interaction is strongest in the MGE. From these studies, and earlier studies, we present a model of the transcriptional network that regulates early steps of subcortical development.
Copyright © 2012 Wiley Periodicals, Inc.
Figures




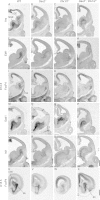
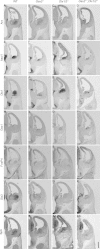
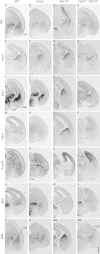


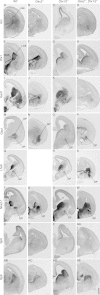







References
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. - PubMed
-
- Casarosa S, Fode C, Guillemot F. Mash1 regulates neurogenesis in the ventral telencephalon. Development. 1999;126:525–534. - PubMed
-
- Castro DS, Skowronska-Krawczyk D, Armant O, Donaldson IJ, Parras C, Hunt C, Critchley JA, Nguyen L, Gossler A, Gottgens B, Matter JM, Guillemot F. Proneural bHLH and Brn proteins coregulate a neurogenic program through cooperative binding to a conserved DNA motif. Dev Cell. 2006;11:831–844. - PubMed
-
- Cobos I, Calcagnotto ME, Vilaythong AJ, Thwin MT, Noebels JL, Baraban SC, Rubenstein JL. Mice lacking Dlx1 show subtype-specific loss of interneurons, reduced inhibition and epilepsy. Nat Neurosci. 2005a;8:1059–1068. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

