Autophagy and selective deployment of Atg proteins in antiviral defense
- PMID: 23042773
- PMCID: PMC3534236
- DOI: 10.1093/intimm/dxs101
Autophagy and selective deployment of Atg proteins in antiviral defense
Abstract
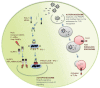
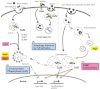
Autophagy is an evolutionarily ancient process eukaryotic cells utilize to remove and recycle intracellular material in order to maintain cellular homeostasis. In metazoans, the autophagy machinery not only functions in this capacity but also has evolved to perform a diverse repertoire of intracellular transport and regulatory functions. In response to virus infections, the autophagy machinery degrades viruses, shuttles viral pathogen-associated molecular patterns to endosomes containing Toll-like receptors, facilitates viral-antigen processing for major histocompatibility complex presentation and transports antiviral proteins to viral replication sites. This is accomplished through canonical autophagy or through processes involving distinct subsets of the autophagy-related genes (Atgs). Herein, we discuss how the variable components of the autophagy machinery contribute to antiviral defense and highlight three emerging themes: first, autophagy delivers viral cytosolic components to several distinct endolysosomal compartments; second, Atg proteins act alone, as subgroups or collectively; and third, the specificity of autophagy and the autophagy machinery is achieved by recognition of triggers and selective targeting by adaptors.
Figures

References
-
- Levine B., Klionsky D. J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463 - PubMed
-
- Mizushima N. 2007. Autophagy: process and function. Genes Dev. 21: 2861 - PubMed
-
- Levine B., Klionsky D. J. 2004. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell 6: 463 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- AI064705/AI/NIAID NIH HHS/United States
- AI054359/AI/NIAID NIH HHS/United States
- R01 AI064705/AI/NIAID NIH HHS/United States
- R01 AI081884/AI/NIAID NIH HHS/United States
- R01 AI054359/AI/NIAID NIH HHS/United States
- T32AI07019/AI/NIAID NIH HHS/United States
- F31 AG039163/AG/NIA NIH HHS/United States
- AI062428/AI/NIAID NIH HHS/United States
- AI081884/AI/NIAID NIH HHS/United States
- T32 AI007019/AI/NIAID NIH HHS/United States
- T32 AI055403/AI/NIAID NIH HHS/United States
- U54AI057160/AI/NIAID NIH HHS/United States
- R56 AI062428/AI/NIAID NIH HHS/United States
- R01 AI062428/AI/NIAID NIH HHS/United States
- U54 AI057160/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical

