Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates
- PMID: 23042990
- PMCID: PMC3497493
- DOI: 10.1128/JB.01428-12
Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates
Abstract
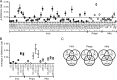
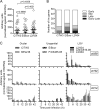
Chlamydia trachomatis is a human bacterial pathogen that multiplies only within an intracellular membrane-bound vacuole, the inclusion. C. trachomatis includes ocular and urogenital strains, usually causing infections restricted to epithelial cells of the conjunctiva and genital mucosa, respectively, and lymphogranuloma venereum (LGV) strains, which can infect macrophages and spread into lymph nodes. However, C. trachomatis genomes display >98% identity at the DNA level. In this work, we studied whether C. trachomatis Inc proteins, which have a bilobed hydrophobic domain that may mediate their insertion in the inclusion membrane, could be a factor determining these different types of infection and tropisms. Analyses of polymorphisms and phylogeny of 48 Inc proteins from 51 strains encompassing the three disease groups showed significant amino acid differences that were mainly due to variations between Inc proteins from LGV and ocular or urogenital isolates. Studies of the evolutionary dynamics of inc genes suggested that 10 of them are likely under positive selection and indicated that most nonsilent mutations are LGV specific. Additionally, real-time quantitative PCR analyses in prototype and clinical strains covering the three disease groups identified three inc genes with LGV-specific expression. We determined the transcriptional start sites of these genes and found LGV-specific nucleotides within their promoters. Thus, subtle variations in the amino acids of a subset of Inc proteins and in the expression of inc genes may contribute to the unique tropism and invasiveness of C. trachomatis LGV strains.
Figures



References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources

