LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects
- PMID: 23043101
- PMCID: PMC3516725
- DOI: 10.1074/jbc.M112.383836
LETM proteins play a role in the accumulation of mitochondrially encoded proteins in Arabidopsis thaliana and AtLETM2 displays parent of origin effects
Abstract
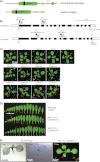
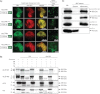
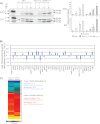
The Arabidopsis thaliana genome contains two genes with homology to the mitochondrial protein LETM1 (leucine zipper-EF-hand-containing transmembrane protein). Inactivation of both genes, Atletm1 and Atletm2, together is lethal. Plants that are hemizygous for AtLETM2 and homozygous for Atletm1 (letm1(-/-) LETM2(+/-)) displayed a mild retarded growth phenotype during early seedling growth. It was shown that accumulation of mitochondrial proteins was reduced in hemizygous (letm1(-/-) LETM2(+/-)) plants. Examination of respiratory chain proteins by Western blotting, blue native PAGE, and enzymatic activity assays revealed that the steady state level of ATP synthase was reduced in abundance, whereas the steady state levels of other respiratory chain proteins remained unchanged. The absence of a functional maternal AtLETM2 allele in an Atletm1 mutant background resulted in early seed abortion. Reciprocal crosses revealed that maternally, but not paternally, derived AtLETM2 was absolutely required for seed development. This requirement for a functional maternal allele of AtLETM2 was confirmed using direct sequencing of reciprocal crosses of Col-0 and Ler accessions. Furthermore, AtLETM2 promoter β-glucuronidase constructs displayed exclusive maternal expression patterns.
Figures




References
-
- Dyall S. D., Brown M. T., Johnson P. J. (2004) Ancient invasions. From endosymbionts to organelles. Science 304, 253–257 - PubMed
-
- Burger G., Gray M. W., Lang B. F. (2003) Mitochondrial genomes. Anything goes. Trends Genet. 19, 709–716 - PubMed
-
- Gray M. W., Burger G., Lang B. F. (1999) Mitochondrial evolution. Science 283, 1476–1481 - PubMed
-
- Gray M. W., Lang B. F., Burger G. (2004) Mitochondria of protists. Annu. Rev. Genet. 38, 477–524 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

