ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs
- PMID: 23043107
- PMCID: PMC3501051
- DOI: 10.1074/jbc.M112.378109
ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs
Abstract
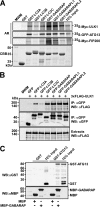
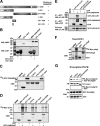
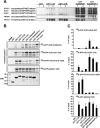
Autophagy is a lysosome-dependent degradation system conserved among eukaryotes. The mammalian Atg1 homologues, Unc-51 like kinase (ULK) 1 and 2, are multifunctional proteins with roles in autophagy, neurite outgrowth, and vesicle transport. The mammalian ULK complex involved in autophagy consists of ULK1, ULK2, ATG13, FIP200, and ATG101. We have used pulldown and peptide array overlay assays to study interactions between the ULK complex and six different ATG8 family proteins. Strikingly, in addition to ULK1 and ULK2, ATG13 and FIP200 interacted with human ATG8 proteins, all with strong preference for the GABARAP subfamily. Similarly, yeast and Drosophila Atg1 interacted with their respective Atg8 proteins, demonstrating the evolutionary conservation of the interaction. Use of peptide arrays allowed precise mapping of the functional LIR motifs, and two-dimensional scans of the ULK1 and ATG13 LIR motifs revealed which substitutions that were tolerated. This information, combined with an analysis of known LIR motifs, provides us with a clearer picture of sequence requirements for LIR motifs. In addition to the known requirements of the aromatic and hydrophobic residues of the core motif, we found the interactions to depend strongly on acidic residues surrounding the central core LIR motifs. A preference for either a hydrophobic residue or an acidic residue following the aromatic residue in the LIR motif is also evident. Importantly, the LIR motif is required for starvation-induced association of ULK1 with autophagosomes. Our data suggest that ATG8 proteins act as scaffolds for assembly of the ULK complex at the phagophore.
Figures






References
-
- Tomoda T., Bhatt R. S., Kuroyanagi H., Shirasawa T., Hatten M. E. (1999) A mouse serine/threonine kinase homologous to C. elegans UNC51 functions in parallel fiber formation of cerebellar granule neurons. Neuron 24, 833–846 - PubMed
-
- Young A. R., Chan E. Y., Hu X. W., Köchl R., Crawshaw S. G., High S., Hailey D. W., Lippincott-Schwartz J., Tooze S. A. (2006) Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 119, 3888–3900 - PubMed
-
- Chan E. Y. (2012) Regulation and function of uncoordinated-51 like kinase proteins. Antioxidants Redox Signal. 17, 775–785 - PubMed
-
- Weidberg H., Shvets E., Elazar Z. (2011) Biogenesis and cargo selectivity of autophagosomes. Annu. Rev. Biochem. 80, 125–156 - PubMed
-
- Nakatogawa H., Suzuki K., Kamada Y., Ohsumi Y. (2009) Dynamics and diversity in autophagy mechanisms. Lessons from yeast. Nat. Rev. Mol. Cell Biol. 10, 458–467 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

