Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations
- PMID: 23043116
- PMCID: PMC3491530
- DOI: 10.1073/pnas.1202997109
Phylogenetic modeling of lateral gene transfer reconstructs the pattern and relative timing of speciations
Abstract
The timing of the evolution of microbial life has largely remained elusive due to the scarcity of prokaryotic fossil record and the confounding effects of the exchange of genes among possibly distant species. The history of gene transfer events, however, is not a series of individual oddities; it records which lineages were concurrent and thus provides information on the timing of species diversification. Here, we use a probabilistic model of genome evolution that accounts for differences between gene phylogenies and the species tree as series of duplication, transfer, and loss events to reconstruct chronologically ordered species phylogenies. Using simulations we show that we can robustly recover accurate chronologically ordered species phylogenies in the presence of gene tree reconstruction errors and realistic rates of duplication, transfer, and loss. Using genomic data we demonstrate that we can infer rooted species phylogenies using homologous gene families from complete genomes of 10 bacterial and archaeal groups. Focusing on cyanobacteria, distinguished among prokaryotes by a relative abundance of fossils, we infer the maximum likelihood chronologically ordered species phylogeny based on 36 genomes with 8,332 homologous gene families. We find the order of speciation events to be in full agreement with the fossil record and the inferred phylogeny of cyanobacteria to be consistent with the phylogeny recovered from established phylogenomics methods. Our results demonstrate that lateral gene transfers, detected by probabilistic models of genome evolution, can be used as a source of information on the timing of evolution, providing a valuable complement to the limited prokaryotic fossil record.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
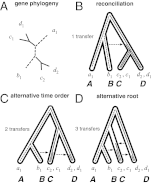


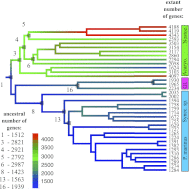
References
-
- Benton MJ, Ayala FJ. Dating the tree of life. Science. 2003;300:1698–1700. - PubMed
-
- Knoll AH. Life on a Young Planet: The First Three Billion Years of Evolution on Earth. Princeton, NJ: Princeton Univ Press; 2003.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

