6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2
- PMID: 23045360
- PMCID: PMC3531295
- DOI: 10.1093/glycob/cws140
6-alkynyl fucose is a bioorthogonal analog for O-fucosylation of epidermal growth factor-like repeats and thrombospondin type-1 repeats by protein O-fucosyltransferases 1 and 2
Abstract
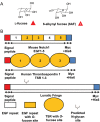
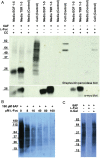
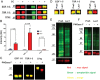
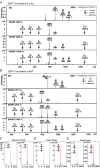
Protein O-fucosyltransferase 1 (Pofut1) and protein O-fucosyltransferase 2 (Pofut2) add O-linked fucose at distinct consensus sequences in properly folded epidermal growth factor (EGF)-like repeats and thrombospondin type-1 (TSR) repeats, respectively. Glycan chain elongation past O-fucose can occur to yield a tetrasaccharide on EGF repeats and a disaccharide on TSRs. Elimination of Pofut1 in mice causes embryonic lethality with Notch-like phenotypes demonstrating that O-fucosylation of Notch is essential for its function. Similarly, elimination of Pofut2 results in an early embryonic lethal phenotype in mice, although the molecular mechanism for the lethality is unknown. The recent development of sugar analogs has revolutionized the study of glycans by providing a convenient method for labeling and tracking glycosylation. In order to study O-fucosylation, we took advantage of the recently developed reporter, 6-alkynyl fucose. Using the Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC), or "click" reaction, azido-biotin allows tagging and detection of 6AF-modified proteins. Here we examine whether proteins containing EGF repeats or TSRs with O-fucose consensus sequences are specifically modified with 6AF in cell culture. Using mass spectrometry (MS), we demonstrate that 6AF is efficiently incorporated onto the appropriate consensus sequences on EGF repeats and TSRs. Furthermore, the elongation of the O-fucose monosaccharide on EGF repeats and TSRs is not hampered when 6AF is used. These results show that 6AF is efficiently utilized in a truly bioorthogonal manner by Pofut1, Pofut2 and the enzymes that elongate O-fucose, providing evidence that 6AF is a significant new tool in the study of protein O-fucosylation.
Figures

References
-
- Becker DJ, Lowe JB. Fucose: Biosynthesis and biological function in mammals. Glycobiology. 2003;13:41R–53R. doi:10.1093/glycob/cwg054. - DOI - PubMed
-
- Besanceney-Webler C, Jiang H, Wang W, Baughn AD, Wu P. Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorg Med Chem Lett. 2011;21:4989–4992. doi:10.1016/j.bmcl.2011.05.038. - DOI - PMC - PubMed
-
- Bruckner K, Perez L, Clausen H, Cohen S. Glycosyltransferase activity of Fringe modulates Notch-Delta interactions. Nature. 2000;406:411–415. doi:10.1038/35019075. - DOI - PubMed
-
- Dehnert KW, Baskin JM, Laughlin ST, Beahm BJ, Naidu NN, Amacher SL, Bertozzi CR. Imaging the sialome during zebrafish development with copper-free click chemistry. Chembiochem. 2012;13:353–357. doi:10.1002/cbic.201100649. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

