Extensive evolutionary and functional diversity among mammalian AIM2-like receptors
- PMID: 23045604
- PMCID: PMC3478938
- DOI: 10.1084/jem.20121960
Extensive evolutionary and functional diversity among mammalian AIM2-like receptors
Abstract
Innate immune detection of nucleic acids is important for initiation of antiviral responses. Detection of intracellular DNA activates STING-dependent type I interferons (IFNs) and the ASC-dependent inflammasome. Certain members of the AIM2-like receptor (ALR) gene family contribute to each of these pathways, but most ALRs remain uncharacterized. Here, we identify five novel murine ALRs and perform a phylogenetic analysis of mammalian ALRs, revealing a remarkable diversification of these receptors among mammals. We characterize the expression, localization, and functions of the murine and human ALRs and identify novel activators of STING-dependent IFNs and the ASC-dependent inflammasome. These findings validate ALRs as key activators of the antiviral response and provide an evolutionary and functional framework for understanding their roles in innate immunity.
Figures


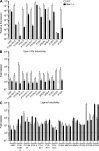
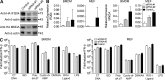

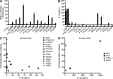
References
-
- Bürckstümmer T., Baumann C., Blüml S., Dixit E., Dürnberger G., Jahn H., Planyavsky M., Bilban M., Colinge J., Bennett K.L., Superti-Furga G. 2009. An orthogonal proteomic-genomic screen identifies AIM2 as a cytoplasmic DNA sensor for the inflammasome. Nat. Immunol. 10:266–272 10.1038/ni.1702 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

