B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1
- PMID: 23045607
- PMCID: PMC3478936
- DOI: 10.1084/jem.20111504
B and T cells collaborate in antiviral responses via IL-6, IL-21, and transcriptional activator and coactivator, Oct2 and OBF-1
Abstract
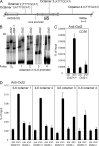
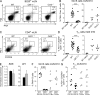
A strong humoral response to infection requires the collaboration of several hematopoietic cell types that communicate via antigen presentation, surface coreceptors and their ligands, and secreted factors. The proinflammatory cytokine IL-6 has been shown to promote the differentiation of activated CD4(+) T cells into T follicular helper cells (T(FH) cells) during an immune response. T(FH) cells collaborate with B cells in the formation of germinal centers (GCs) during T cell-dependent antibody responses, in part through secretion of critical cytokines such as IL-21. In this study, we demonstrate that loss of either IL-6 or IL-21 has marginal effects on the generation of T(FH) cells and on the formation of GCs during the response to acute viral infection. However, mice lacking both IL-6 and IL-21 were unable to generate a robust T(FH) cell-dependent immune response. We found that IL-6 production in follicular B cells in the draining lymph node was an important early event during the antiviral response and that B cell-derived IL-6 was necessary and sufficient to induce IL-21 from CD4(+) T cells in vitro and to support T(FH) cell development in vivo. Finally, the transcriptional activator Oct2 and its cofactor OBF-1 were identified as regulators of Il6 expression in B cells.
Figures




References
-
- Baccam M., Woo S.Y., Vinson C., Bishop G.A. 2003. CD40-mediated transcriptional regulation of the IL-6 gene in B lymphocytes: involvement of NF-kappa B, AP-1, and C/EBP. J. Immunol. 170:3099–3108 - PubMed
-
- Batten M., Ramamoorthi N., Kljavin N.M., Ma C.S., Cox J.H., Dengler H.S., Danilenko D.M., Caplazi P., Wong M., Fulcher D.A., et al. 2010. IL-27 supports germinal center function by enhancing IL-21 production and the function of T follicular helper cells. J. Exp. Med. 207:2895–2906 10.1084/jem.20100064 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

