Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA
- PMID: 23046130
- PMCID: PMC3479221
- DOI: 10.1111/imr.12005
Structural insights into key sites of vulnerability on HIV-1 Env and influenza HA
Abstract
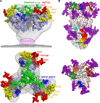
Human immunodeficiency virus-1 (HIV-1) envelope protein (Env) and influenza hemagglutinin (HA) are the surface glycoproteins responsible for viral entry into host cells, the first step in the virus life cycle necessary to initiate infection. These glycoproteins exhibit a high degree of sequence variability and glycosylation, which are used as strategies to escape host immune responses. Nonetheless, antibodies with broadly neutralizing activity against these viruses have been isolated that have managed to overcome these barriers. Here, we review recent advances in the structural characterization of these antibodies with their viral antigens that defines a few sites of vulnerability on these viral spikes. These broadly neutralizing antibodies tend to focus their recognition on the sites of similar function between the two viruses: the receptor-binding site and membrane fusion machinery. However, some sites of recognition are unique to the virus neutralized, such as the dense shield of oligomannose carbohydrates on HIV-1 Env. These observations are discussed in the context of structure-based design strategies to aid in vaccine design or development of antivirals.
© 2012 John Wiley & Sons A/S.
Conflict of interest statement
The authors state to have no financial or personal relationships that could be viewed as a potential conflict of interest.
Figures


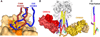
References
-
- Gray RH, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. Lancet. 2001;357:1149–1153. - PubMed
-
- Forsman A, Weiss RA. Why is HIV a pathogen? Trends Microbiol. 2008;16:555–560. - PubMed
-
- Gelderblom HR, Hausmann EH, Ozel M, Pauli G, Koch MA. Fine structure of human immunodeficiency virus (HIV) and immunolocalization of structural proteins. Virology. 1987;156:171–176. - PubMed
-
- Zhu P, et al. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature. 2006;441:847–852. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

