Ubiquitin receptors and protein quality control
- PMID: 23046644
- PMCID: PMC3571097
- DOI: 10.1016/j.yjmcc.2012.09.012
Ubiquitin receptors and protein quality control
Abstract
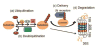
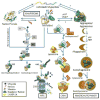
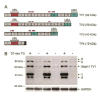
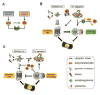
Protein quality control (PQC) is essential to intracellular proteostasis and is carried out by sophisticated collaboration between molecular chaperones and targeted protein degradation. The latter is performed by proteasome-mediated degradation, chaperone-mediated autophagy (CMA), and selective macroautophagy, and collectively serves as the final line of defense of PQC. Ubiquitination and subsequently ubiquitin (Ub) receptor proteins (e.g., p62 and ubiquilins) are important common factors for targeting misfolded proteins to multiple quality control destinies, including the proteasome, lysosomes, and perhaps aggresomes, as well as for triggering mitophagy to remove defective mitochondria. PQC inadequacy, particularly proteasome functional insufficiency, has been shown to participate in cardiac pathogenesis. Tremendous advances have been made in unveiling the changes of PQC in cardiac diseases. However, the investigation into the molecular pathways regulating PQC in cardiac (patho)physiology, including the function of most ubiquitin receptor proteins in the heart, has only recently been initiated. A better understanding of molecular mechanisms governing PQC in cardiac physiology and pathology will undoubtedly provide new insights into cardiac pathogenesis and promote the search for novel therapeutic strategies to more effectively battle heart disease.This article is part of a Special Issue entitled "Focus on Cardiac Metabolism".
Copyright © 2012. Published by Elsevier Ltd.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

