Spatiotemporal regulation of the ubiquitinated cargo-binding activity of Rabex-5 in the endocytic pathway
- PMID: 23048039
- PMCID: PMC3504772
- DOI: 10.1074/jbc.M112.411793
Spatiotemporal regulation of the ubiquitinated cargo-binding activity of Rabex-5 in the endocytic pathway
Abstract
Background: The regulatory mechanism underlying the interaction of the Rabex-5 MIU domain with ubiquitinated cargos remains unclear.
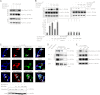
Results: Rabex-5 guanine nucleotide exchange factor (GEF) mutants affected interactions of ubiquitinated cargos.
Conclusion: GDP/GTP exchange in the GEF domain controls the MIU domain interactions with the ubiquitinated cargos.
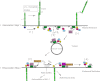
Significance: Rabex-5 GEF activity acts as an intramolecular switch for spatiotemporal trafficking of the ubiquitinated cargos. Ubiquitin (Ub)-dependent endocytosis of membrane proteins requires precise molecular recognition of ubiquitinated cargo by Ub-binding proteins (UBPs). Many UBPs are often themselves monoubiquitinated, a mechanism referred to as coupled monoubiquitination, which prevents them from binding in trans to the ubiquitinated cargo. However, the spatiotemporal regulatory mechanism underlying the interaction of UBPs with the ubiquitinated cargo, via their Ub-binding domains (UBDs) remains unclear. Previously, we reported the interaction of Rabex-5, a UBP and guanine nucleotide exchange factor (GEF) for Rab5, with ubiquitinated neural cell adhesion molecule L1, via its motif interacting with Ub (MIU) domain. This interaction is critical for the internalization and sorting of the ubiquitinated L1 into endosomal/lysosomal compartments. The present study demonstrated that the interaction of Rabex-5 with Rab5 depends specifically on interaction of the MIU domain with the ubiquitinated L1 to drive its internalization. Notably, impaired GEF mutants and the Rabex-5(E213A) mutant increased the flexibility of the hinge region in the HB-VPS9 tandem domain, which significantly affected their interactions with the ubiquitinated L1. In addition, GEF mutants increased the catalytic efficiency, which resulted in a reduced interaction with the ubiquitinated L1. Furthermore, the coupled monoubiquitination status of Rabex-5 was found to be significantly associated with interaction of Rabex-5 and the ubiquitinated L1. Collectively, our study reveals a novel mechanism, wherein the GEF activity of Rabex-5 acts as an intramolecular switch orchestrating ubiquitinated cargo-binding activity and coupled monoubiquitination to permit the spatiotemporal dynamic exchange of the ubiquitinated cargos.
Figures





References
-
- Mukhopadhyay D., Riezman H. (2007) Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science 315, 201–205 - PubMed
-
- Raiborg C., Stenmark H. (2009) The ESCRT machinery in endosomal sorting of ubiquitylated membrane proteins. Nature 458, 445–452 - PubMed
-
- Hicke L., Schubert H. L., Hill C. P. (2005) Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6, 610–621 - PubMed
-
- Traub L. M., Lukacs G. L. (2007) Decoding ubiquitin sorting signals for clathrin-dependent endocytosis by CLASPs. J. Cell Sci. 120, 543–553 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

