Combinatorial regulation of tissue specification by GATA and FOG factors
- PMID: 23048181
- PMCID: PMC3472596
- DOI: 10.1242/dev.080440
Combinatorial regulation of tissue specification by GATA and FOG factors
Abstract
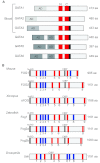
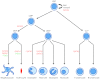
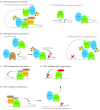
The development of complex organisms requires the formation of diverse cell types from common stem and progenitor cells. GATA family transcriptional regulators and their dedicated co-factors, termed Friend of GATA (FOG) proteins, control cell fate and differentiation in multiple tissue types from Drosophila to man. FOGs can both facilitate and antagonize GATA factor transcriptional regulation depending on the factor, cell, and even the specific gene target. In this review, we highlight recent studies that have elucidated mechanisms by which FOGs regulate GATA factor function and discuss how these factors use these diverse modes of gene regulation to control cell lineage specification throughout metazoans.
Figures
References
-
- Abel T., Michelson A. M., Maniatis T. (1993). A Drosophila GATA family member that binds to Adh regulatory sequences is expressed in the developing fat body. Development 119, 623-633 - PubMed
-
- Anttonen M., Ketola I., Parviainen H., Pusa A.-K., Heikinheimo M. (2003). FOG-2 and GATA-4 are coexpressed in the mouse ovary and can modulate mullerian-inhibiting substance expression. Biol. Reprod. 68, 1333-1340 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

