Probiotics, prebiotics, and synbiotics: gut and beyond
- PMID: 23049548
- PMCID: PMC3459241
- DOI: 10.1155/2012/872716
Probiotics, prebiotics, and synbiotics: gut and beyond
Abstract
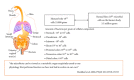
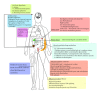
The human intestinal tract has been colonized by thousands of species of bacteria during the coevolution of man and microbes. Gut-borne microbes outnumber the total number of body tissue cells by a factor of ten. Recent metagenomic analysis of the human gut microbiota has revealed the presence of some 3.3 million genes, as compared to the mere 23 thousand genes present in the cells of the tissues in the entire human body. Evidence for various beneficial roles of the intestinal microbiota in human health and disease is expanding rapidly. Perturbation of the intestinal microbiota may lead to chronic diseases such as autoimmune diseases, colon cancers, gastric ulcers, cardiovascular disease, functional bowel diseases, and obesity. Restoration of the gut microbiota may be difficult to accomplish, but the use of probiotics has led to promising results in a large number of well-designed (clinical) studies. Microbiomics has spurred a dramatic increase in scientific, industrial, and public interest in probiotics and prebiotics as possible agents for gut microbiota management and control. Genomics and bioinformatics tools may allow us to establish mechanistic relationships among gut microbiota, health status, and the effects of drugs in the individual. This will hopefully provide perspectives for personalized gut microbiota management.
Figures

References
-
- Kunz C, Kuntz S, Rudloff S. Intestinal flora. Advances in Experimental Medicine and Biology. 2009;639:67–79. - PubMed
-
- Morelli L. Postnatal development of intestinal microflora as influenced by infant nutrition. Journal of Nutrition. 2008;138(supplement 9):S1791–S1795. - PubMed
-
- Ley RE, Peterson DA, Gordon JI. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell. 2006;124(4):837–848. - PubMed
-
- Savage DC. Associations of indigenous microorganisms with gastrointestinal mucosal epithelia. American Journal of Clinical Nutrition. 1970;23(11):1495–1501. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

