A novel group of Moraxella catarrhalis UspA proteins mediates cellular adhesion via CEACAMs and vitronectin
- PMID: 23049802
- PMCID: PMC3458076
- DOI: 10.1371/journal.pone.0045452
A novel group of Moraxella catarrhalis UspA proteins mediates cellular adhesion via CEACAMs and vitronectin
Abstract
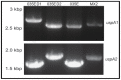
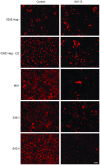
Moraxella catarrhalis (Mx) is a common cause of otitis media and exacerbation of chronic obstructive pulmonary disease, an increasing worldwide problem. Surface proteins UspA1 and UspA2 of Mx bind to a number of human receptors and may function in pathogenesis. Genetic recombination events in the pathogen can generate hybrid proteins termed UspA2H. However, whether certain key functions (e.g. UspA1-specific CEACAM binding) can be exchanged between these adhesin families remains unknown. In this study, we have shown that Mx can incorporate the UspA1 CEACAM1-binding region not only into rare UspA1 proteins devoid of CEACAM-binding ability, but also into UspA2 which normally lack this capacity. Further, a screen of Mx isolates revealed the presence of novel UspA2 Variant proteins (UspA2V) in ∼14% of the CEACAM-binding population. We demonstrate that the expression of UspA2/2V with the CEACAM-binding domain enable Mx to bind both to cell surface CEACAMs and to integrins, the latter via vitronectin. Such properties of UspA2/2V have not been reported to date. The studies demonstrate that the UspA family is much more heterogeneous than previously believed and illustrate the in vivo potential for exchange of functional regions between UspA proteins which could convey novel adhesive functions whilst enhancing immune evasion.
Conflict of interest statement
Figures




References
-
- Murphy TF, Parameswaran GI (2009) Moraxella catarrhalis, a human respiratory tract pathogen. Clin Infect Dis 49: 124–131. - PubMed
-
- Cripps AW, Otczyk DC, Kyd JM (2005) Bacterial otitis media: a vaccine preventable disease? Vaccine 23: 2304–2310. - PubMed
-
- Mannino DM, Buist AS (2007) Global burden of COPD: risk factors, prevalence, and future trends. Lancet 370: 765–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

