A systematic investigation of differential effects of cell culture substrates on the extent of artifacts in single-molecule tracking
- PMID: 23049831
- PMCID: PMC3458086
- DOI: 10.1371/journal.pone.0045655
A systematic investigation of differential effects of cell culture substrates on the extent of artifacts in single-molecule tracking
Abstract
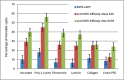
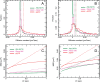
Single-molecule techniques are being increasingly applied to biomedical investigation, notwithstanding the numerous challenges they pose in terms of signal-to-noise ratio issues. Non-specific binding of probes to glass substrates, in particular, can produce experimental artifacts due to spurious molecules on glass, which can be particularly deleterious in live-cell tracking experiments. In order to resolve the issue of non-specific probe binding to substrates, we performed systematic testing of a range of available surface coatings, using three different proteins, and then extended our assessment to the ability of these coatings to foster cell growth and retain non-adhesive properties. Linear PEG, a passivating agent commonly used both in immobilized-molecule single-molecule techniques and in tissue engineering, is able to both successfully repel non-specific adhesion of fluorescent probes and to foster cell growth when functionalized with appropriate adhesive peptides. Linear PEG treatment results in a significant reduction of tracking artifacts in EGFR tracking with Affibody ligands on a cell line expressing EGFR-eGFP. The findings reported herein could be beneficial to a large number of experimental situations where single-molecule or single-particle precision is required.
Conflict of interest statement
Figures

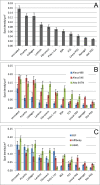

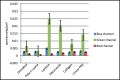
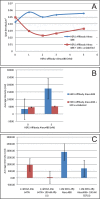

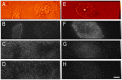
References
-
- Selvin PR, Ha T (2007) Single-Molecule Techniques: A Laboratory Manual: Cold Spring Harbor Laboratory Press. 507 p.
-
- Joo C, Balci H, Ishitsuka Y, Buranachai C, Ha T (2008) Advances in single-molecule fluorescence methods for molecular biology. Annual review of biochemistry 77: 51–76. - PubMed
-
- Yildiz A, Selvin PR (2005) Fluorescence imaging with one nanometer accuracy: application to molecular motors. Accounts of chemical research 38: 574–582. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

