Validation of the Cancer BioChip System as a 3D siRNA screening tool for breast cancer targets
- PMID: 23049944
- PMCID: PMC3458802
- DOI: 10.1371/journal.pone.0046086
Validation of the Cancer BioChip System as a 3D siRNA screening tool for breast cancer targets
Abstract
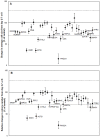
Genomic studies have revealed that breast cancer consists of a complex biological process with patient-specific genetic variations, revealing the need for individualized cancer diagnostic testing and selection of patient-specific optimal therapies. One of the bottlenecks in translation of genomic breakthroughs to the clinic is the lack of functional genomic assays that have high clinical translatability. Anchorage-independent three-dimensional (3D) growth assays are considered to be the gold-standard for chemosensitivity testing, and leads identified with these assays have high probability of clinical success. The Cancer BioChip System (CBCS) allows for the simultaneous, quantitative, and real time evaluation of multitudes of anchorage-independent breast cancer cell growth inhibitors. We employed a Test Cancer BioChip that contains silencing RNAs (siRNAs) targeting cancer-related genes to identify 3D-specific effectors of breast cancer cell growth. We compared the effect of these siRNAs on colony growth of the hormone receptor positive (MCF7) and Human Epidermal Growth Factor Receptor 2/c- Erythroblastic Leukemia Viral Oncogene Homolog 2 (HER2/c-erb-b2) positive (SK-BR-3) cells on the Test Cancer BioChip. Our results confirmed cell-specific inhibition of MCF7 and SK-BR-3 colony formation by estrogen receptor α (ESR1) and (ERBB2) siRNA, respectively. Both cell lines were also suppressed by Phosphoinositide-3-kinase Catalytic, alpha Polypeptide (PIK3CA) siRNA. Interestingly, we have observed responses to siRNA that are unique to this 3D setting. For example, ß-actin (ACTB) siRNA suppressed colony growth in both cell types while Cathepsin L2 (CTSL2) siRNA caused opposite effects. These results further validate the importance of the CBCS as a tool for the identification of clinically relevant breast cancer targets.
Conflict of interest statement
Figures





References
-
- Berns K, Horlings HM, Hennessy BT, Madiredjo M, Hijmans EM, et al. (2007) A functional genetic approach identifies the PI3K pathway as a major determinant of trastuzumab resistance in breast cancer. Cancer Cell 12: 395–402. - PubMed
-
- Iorns E, Lord CJ, Ashworth A (2009) Parallel RNAi and compound screens identify the PDK1 pathway as a target for tamoxifen sensitization. Biochem J 417: 361–370. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

