Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages
- PMID: 23050217
- PMCID: PMC3442828
- DOI: 10.4161/bact.20632
Mobilization of pathogenicity islands by Staphylococcus aureus strain Newman bacteriophages
Abstract
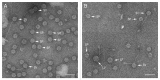
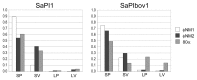
Staphylococcus aureus pathogenicity islands (SaPIs) are mobile genetic elements that encode virulence factors and depend on helper phages for their mobilization. Such mobilization is specific and depends on the ability of a phage protein to inactivate the SaPI repressor Stl. Phage 80α can mobilize several SaPIs, including SaPI1 and SaPIbov1, via its Sri and Dut proteins, respectively. In many cases, the capsids formed in the presence of the SaPI are smaller than those normally produced by the phage. Two SaPI-encoded proteins, CpmA and CpmB, are involved in this size determination process. S. aureus strain Newman contains four prophages, named φNM1 through φNM4. Phages φNM1 and φNM2 are very similar to phage 80α in the structural genes, and encode almost identical Sri proteins, while their Dut proteins are highly divergent. We show that φNM1 and φNM2 are able to mobilize both SaPI1 and SaPIbov1 and yield infectious transducing particles. The majority of the capsids formed in all cases are small, showing that both SaPIs can redirect the capsid size of both φNM1 and φNM2.
Figures



References
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
