Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis
- PMID: 23050870
- PMCID: PMC3542171
- DOI: 10.1186/1471-2164-13-544
Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis
Abstract
Background: The MYB gene family comprises one of the richest groups of transcription factors in plants. Plant MYB proteins are characterized by a highly conserved MYB DNA-binding domain. MYB proteins are classified into four major groups namely, 1R-MYB, 2R-MYB, 3R-MYB and 4R-MYB based on the number and position of MYB repeats. MYB transcription factors are involved in plant development, secondary metabolism, hormone signal transduction, disease resistance and abiotic stress tolerance. A comparative analysis of MYB family genes in rice and Arabidopsis will help reveal the evolution and function of MYB genes in plants.
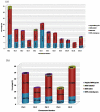
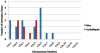
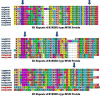
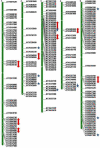
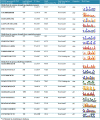
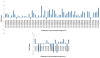
Results: A genome-wide analysis identified at least 155 and 197 MYB genes in rice and Arabidopsis, respectively. Gene structure analysis revealed that MYB family genes possess relatively more number of introns in the middle as compared with C- and N-terminal regions of the predicted genes. Intronless MYB-genes are highly conserved both in rice and Arabidopsis. MYB genes encoding R2R3 repeat MYB proteins retained conserved gene structure with three exons and two introns, whereas genes encoding R1R2R3 repeat containing proteins consist of six exons and five introns. The splicing pattern is similar among R1R2R3 MYB genes in Arabidopsis. In contrast, variation in splicing pattern was observed among R1R2R3 MYB members of rice. Consensus motif analysis of 1kb upstream region (5' to translation initiation codon) of MYB gene ORFs led to the identification of conserved and over-represented cis-motifs in both rice and Arabidopsis. Real-time quantitative RT-PCR analysis showed that several members of MYBs are up-regulated by various abiotic stresses both in rice and Arabidopsis.
Conclusion: A comprehensive genome-wide analysis of chromosomal distribution, tandem repeats and phylogenetic relationship of MYB family genes in rice and Arabidopsis suggested their evolution via duplication. Genome-wide comparative analysis of MYB genes and their expression analysis identified several MYBs with potential role in development and stress response of plants.
Figures


References
-
- Lipsick JS. One billion years of Myb. Oncogene. 1996;13:223–235. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

