New perspectives on viable microbial communities in low-biomass cleanroom environments
- PMID: 23051695
- PMCID: PMC3554398
- DOI: 10.1038/ismej.2012.114
New perspectives on viable microbial communities in low-biomass cleanroom environments
Abstract
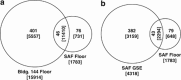
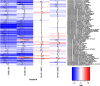
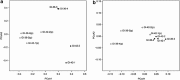
The advent of phylogenetic DNA microarrays and high-throughput pyrosequencing technologies has dramatically increased the resolution and accuracy of detection of distinct microbial lineages in mixed microbial assemblages. Despite an expanding array of approaches for detecting microbes in a given sample, rapid and robust means of assessing the differential viability of these cells, as a function of phylogenetic lineage, remain elusive. In this study, pre-PCR propidium monoazide (PMA) treatment was coupled with downstream pyrosequencing and PhyloChip DNA microarray analyses to better understand the frequency, diversity and distribution of viable bacteria in spacecraft assembly cleanrooms. Sample fractions not treated with PMA, which were indicative of the presence of both live and dead cells, yielded a great abundance of highly diverse bacterial pyrosequences. In contrast, only 1% to 10% of all of the pyrosequencing reads, arising from a few robust bacterial lineages, originated from sample fractions that had been pre-treated with PMA. The results of PhyloChip analyses of PMA-treated and -untreated sample fractions were in agreement with those of pyrosequencing. The viable bacterial population detected in cleanrooms devoid of spacecraft hardware was far more diverse than that observed in cleanrooms that housed mission-critical spacecraft hardware. The latter was dominated by hardy, robust organisms previously reported to survive in oligotrophic cleanroom environments. Presented here are the findings of the first ever comprehensive effort to assess the viability of cells in low-biomass environmental samples, and correlate differential viability with phylogenetic affiliation.
Figures