Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress
- PMID: 23052213
- PMCID: PMC11113399
- DOI: 10.1007/s00018-012-1173-4
Stress-induced self-cannibalism: on the regulation of autophagy by endoplasmic reticulum stress
Abstract
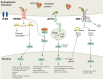
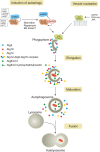
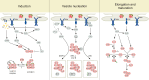
Macroautophagy (autophagy) is a cellular catabolic process which can be described as a self-cannibalism. It serves as an essential protective response during conditions of endoplasmic reticulum (ER) stress through the bulk removal and degradation of unfolded proteins and damaged organelles; in particular, mitochondria (mitophagy) and ER (reticulophagy). Autophagy is genetically regulated and the autophagic machinery facilitates removal of damaged cell components and proteins; however, if the cell stress is acute or irreversible, cell death ensues. Despite these advances in the field, very little is known about how autophagy is initiated and how the autophagy machinery is transcriptionally regulated in response to ER stress. Some three dozen autophagy genes have been shown to be required for the correct assembly and function of the autophagic machinery; however; very little is known about how these genes are regulated by cellular stress. Here, we will review current knowledge regarding how ER stress and the unfolded protein response (UPR) induce autophagy, including description of the different autophagy-related genes which are regulated by the UPR.
Figures
References
-
- Gorman AM, Healy SJ, Jager R, Samali A (2012) Stress management at the ER: regulators of ER stress-induced apoptosis. Pharmacol Ther. doi:10.1016/j.pharmthera.2012.02.003 - PubMed
-
- Cawley K, Deegan S, Samali A, Gupta S (2011) In: Michael Conn P (ed) Methods in enzymology, vol 490. Academic, London, p 31–51 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

