Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions
- PMID: 23052255
- PMCID: PMC4234119
- DOI: 10.1158/1078-0432.CCR-12-0838
Identification of KIF5B-RET and GOPC-ROS1 fusions in lung adenocarcinomas through a comprehensive mRNA-based screen for tyrosine kinase fusions
Abstract
Background: The mutually exclusive pattern of the major driver oncogenes in lung cancer suggests that other mutually exclusive oncogenes exist. We conducted a systematic search for tyrosine kinase fusions by screening all tyrosine kinases for aberrantly high RNA expression levels of the 3' kinase domain (KD) exons relative to more 5' exons.
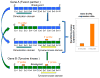
Methods: We studied 69 patients (including five never smokers and 64 current or former smokers) with lung adenocarcinoma negative for all major mutations in KRAS, EGFR, BRAF, MEK1, HER2, and for ALK fusions (termed "pan-negative"). A NanoString-based assay was designed to query the transcripts of 90 tyrosine kinases at two points: 5' to the KD and within the KD or 3' to it. Tumor RNAs were hybridized to the NanoString probes and analyzed for outlier 3' to 5' expression ratios. Presumed novel fusion events were studied by rapid amplification of cDNA ends (RACE) and confirmatory reverse transcriptase PCR (RT-PCR) and FISH.
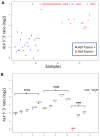
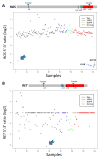
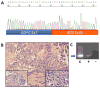
Results: We identified one case each of aberrant 3' to 5' ratios in ROS1 and RET. RACE isolated a GOPC-ROS1 (FIG-ROS1) fusion in the former and a KIF5B-RET fusion in the latter, both confirmed by RT-PCR. The RET rearrangement was also confirmed by FISH. The KIF5B-RET patient was one of only five never smokers in this cohort.
Conclusion: The KIF5B-RET fusion defines an additional subset of lung cancer with a potentially targetable driver oncogene enriched in never smokers with "pan-negative" lung adenocarcinomas. We also report in lung cancer the GOPC-ROS1 fusion originally discovered and characterized in a glioma cell line.
©2012 AACR.
Conflict of interest statement
Figures


References
-
- Pao W, Iafrate AJ, Su Z. Genetically informed lung cancer medicine. J Pathol. 2011;223:230–40. - PubMed
-
- Pao W, Girard N. New driver mutations in non-small-cell lung cancer. Lancet Oncol. 2011;12:175–80. - PubMed
-
- Johnson BE, Kris MG, Kwiatkowski DJ, Iafrate AJ, Wistuba II, Shyr Y, et al. Clinical characteristics of planned 1000 patients with adenocarcinoma of lung (ACL) undergoing genomic characterization in the US Lung Cancer Mutation Consortium (LCMC) J Thorac Oncol. 2011;6:S346.
-
- Arcila ME, Lau C, Jhanwar SC, Zakowski MF, Kris MG, Ladanyi M. Comprehensive analysis for clinically relevant oncogenic driver mutations in 1131 consecutive lung adenocarcinomas. Mod Pathol. 2011;24s1:404A.
-
- Kris MG, Arcila M, Lau C, Rekhtman N, Brzostowski E, Pilloff M, et al. Two year results of LC-MAP: an institutional program to routinely profile tumor specimens for the presence of mutations in targetable pathways in all patients with non-small cell lung cancers. J Thorac Oncol. 2011;6:S347.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

