Identity-by-descent-based heritability analysis in the Northern Finland Birth Cohort
- PMID: 23052944
- PMCID: PMC3543768
- DOI: 10.1007/s00439-012-1230-y
Identity-by-descent-based heritability analysis in the Northern Finland Birth Cohort
Erratum in
- Hum Genet. 2013 Aug;132(8):957-8
Abstract
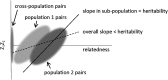
For most complex traits, only a small proportion of heritability is explained by statistically significant associations from genome-wide association studies (GWAS). In order to determine how much heritability can potentially be explained through larger GWAS, several different approaches for estimating total narrow-sense heritability from GWAS data have recently been proposed. These methods include variance components with relatedness estimates from allele-sharing, variance components with relatedness estimates from identity-by-descent (IBD) segments, and regression of phenotypic correlation on relatedness estimates from IBD segments. These methods have not previously been compared on real or simulated data. We analyze the narrow-sense heritability of nine metabolic traits in the Northern Finland Birth Cohort (NFBC) using these methods. We find substantial estimated heritability for several traits, including LDL cholesterol (54 % heritability), HDL cholesterol (46 % heritability), and fasting glucose levels (39 % heritability). Estimates of heritability from the regression-based approach are much lower than variance component estimates in these data, which may be due to the presence of strong population structure. We also investigate the accuracy of the competing approaches using simulated phenotypes based on genotype data from the NFBC. The simulation results substantiate the downward bias of the regression-based approach in the presence of population structure.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

